Gene
KWMTBOMO05844
Pre Gene Modal
BGIBMGA006900
Annotation
PREDICTED:_uncharacterized_protein_LOC101744306_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.52 PlasmaMembrane Reliability : 1.375
Sequence
CDS
ATGGCCAGCAGTTTATACAATATATCAACGTACAGCCGTTTAAATTTATCCGAAAGCGAATCTGATATACCCGTTTTCGTAGACGAATATAGAAAACGCATAAAAAGGCAATTGGCACGAGAAGAGGAGCCGCCGAAGTCTTCGTTTATTTCGTTTGTTCAGAAAGGGCTCAGTATAACCGACCTAACGTGGTTACAAGACAATCGAGATAGATTCTTCAAGGGCACGGAAAACAAGACATTGGTGAAACTGGACTATTTATCAGAATGTGGTTATATTTTTTGTATGGAGTTTTCGCCGGAGGGAGATTTCGTGATTATCGGTCATTCGTCTGGATTAATTCAGATGCGCCACGGTAGTACAGGTACAGTGCTATGTACATTGAGAAATGTGCAGTTTCCGCCCAAGCCAGTGTACGCCATTGAATACAGCATGGTCGAAGAAAGAGTTTGCTACGCTGCTTGTATGGACGGTGCAATTTACAGGATCGAAATACCCAACGTTGTGGCGCCAATAGAGGAACCTCCACTATTCTGTATTAGAGCCGATCCTGCTCTAGAATCGTTCAACATGCAATTCTATGGAAGTCCTGGGATATCATCTTATTCCACACCATTCATCACCCAGCGATCCCCTGCGCTTTCTCTCGGTCTGACCGCTGATCAAACTAAAATGGCGGTCGGTTACGGAGATAGTTCAATAAAAGTATACGATATGGAGACTCAAGAAACAGAGCAGACTTACAAAGTCCACAAACTCCGTCTTCAGTTTATACCGAAGAAGTTACAGAGAATGCACTTCGGTCAAGTTTGCGCTTTGAGATGCCACAAGGATAGGCCCTACATGTTCGCGAGTGCTGCCTGGGACAAAACTTTGAGAATTTGGGACATAAGGTGTCAAGTCGGCTGCGTTATGACGTTCGAAGGTGTTGACGTATGCAGTGACAGCATCGATCTAAATAGGGACCACTGCATAGCCGGCAGCTGGCAAGCGACGGAAGCGTTGACAGTGTGGGATTTAGTAGCACGGAAGAGAATGAATGTAATAAGGGTGCAGAATCGTCGACAAGACGTGGACGGCGAGTACATTTACGCGTGTAGATACTGGAATTCTTCGGAGTACAATCGTAAAGGGAAATATGCCATTGTCGGTGGCAGCGGAACGAATTGCCTGGAAGTGATCAATCTACACAACCGATATATTTCATGTTCTTATCCAGCCTCCGGCACTGTCCTGGCTGTAGCTAGCCACGGTGAGAGGGTAGCCTTCGGTGGAACTGCCCCAGTGTTTAACATTGCGAGTTTCCATGATCCTAAGCACGAGAAGCAAACATATCACGAAGAAGAAATTGATATCGAATACAAGGAAAGTATCATATTACGCGACGAAGATTTTATGTGA
Protein
MASSLYNISTYSRLNLSESESDIPVFVDEYRKRIKRQLAREEEPPKSSFISFVQKGLSITDLTWLQDNRDRFFKGTENKTLVKLDYLSECGYIFCMEFSPEGDFVIIGHSSGLIQMRHGSTGTVLCTLRNVQFPPKPVYAIEYSMVEERVCYAACMDGAIYRIEIPNVVAPIEEPPLFCIRADPALESFNMQFYGSPGISSYSTPFITQRSPALSLGLTADQTKMAVGYGDSSIKVYDMETQETEQTYKVHKLRLQFIPKKLQRMHFGQVCALRCHKDRPYMFASAAWDKTLRIWDIRCQVGCVMTFEGVDVCSDSIDLNRDHCIAGSWQATEALTVWDLVARKRMNVIRVQNRRQDVDGEYIYACRYWNSSEYNRKGKYAIVGGSGTNCLEVINLHNRYISCSYPASGTVLAVASHGERVAFGGTAPVFNIASFHDPKHEKQTYHEEEIDIEYKESIILRDEDFM
Summary
Uniprot
A0A3S2M9Z5
A0A194QQV1
A0A194Q872
A0A0L7LMI7
K7JNK0
A0A232FEX3
+ More
A0A195EUA2 F4WP23 A0A195BA04 A0A158NGL2 A0A0J7L9R5 A0A026WP81 A0A1W4X333 A0A1S3I933 A0A1B6G4E3 A0A1L8GCI2 A0A2T7PYH0 A0DLN0 R7TY40 V4A2K7 A0A210PUC3 H3AGS5 A0A1J4KCQ1 W4YHM2 A2EK02 A0CD35 V9KZ61 F0WRI3 A0A024GKW5 A0A024GK81 A0A1S3NFL6 Q23JY8 A0A024GKC3 K3X9H1 G0QQI5 A7RIL3 A0A1V9ZFR5 A0A1B6MP67 K1QI52 T0Q478 A0A2D4CBH1 A0A067CUN1 V9E791 W2YDS0 A0A080ZAM7 W2K9I3 W2I5E0 W2W2M1 W2PL20 A0A024TFX1 A0A024TG12 W4G8H5 F0XVH6 A0BBZ0 A0A078B6P6 A0A0W8CDI1 Q6BGG5 A0A3F2RP10 G4YR99
A0A195EUA2 F4WP23 A0A195BA04 A0A158NGL2 A0A0J7L9R5 A0A026WP81 A0A1W4X333 A0A1S3I933 A0A1B6G4E3 A0A1L8GCI2 A0A2T7PYH0 A0DLN0 R7TY40 V4A2K7 A0A210PUC3 H3AGS5 A0A1J4KCQ1 W4YHM2 A2EK02 A0CD35 V9KZ61 F0WRI3 A0A024GKW5 A0A024GK81 A0A1S3NFL6 Q23JY8 A0A024GKC3 K3X9H1 G0QQI5 A7RIL3 A0A1V9ZFR5 A0A1B6MP67 K1QI52 T0Q478 A0A2D4CBH1 A0A067CUN1 V9E791 W2YDS0 A0A080ZAM7 W2K9I3 W2I5E0 W2W2M1 W2PL20 A0A024TFX1 A0A024TG12 W4G8H5 F0XVH6 A0BBZ0 A0A078B6P6 A0A0W8CDI1 Q6BGG5 A0A3F2RP10 G4YR99
Pubmed
EMBL
RSAL01000003
RVE54823.1
KQ461175
KPJ07872.1
KQ459302
KPJ01737.1
+ More
JTDY01000584 KOB76569.1 NNAY01000341 OXU29070.1 KQ981965 KYN31736.1 GL888243 EGI63954.1 KQ976542 KYM81060.1 ADTU01015125 LBMM01000169 KMR04642.1 KK107139 QOIP01000003 EZA57768.1 RLU24740.1 GECZ01012461 JAS57308.1 CM004474 OCT81553.1 PZQS01000001 PVD38475.1 CT868492 CAK83947.1 AMQN01011614 KB308980 ELT95875.1 KB202619 ESO89175.1 NEDP02005488 OWF40099.1 AFYH01115911 MLAK01000695 OHT07476.1 AAGJ04033126 DS113410 EAY06986.1 CT868062 CAK68702.1 JW871184 AFP03702.1 FR824259 CCA23946.1 CAIX01000156 CCI47185.1 CCI47183.1 GG662673 EAR97055.2 CCI47184.1 GL376595 GL983641 EGR32520.1 DS469512 EDO48733.1 JNBR01000127 OQR96839.1 GEBQ01002230 JAT37747.1 JH823224 EKC21326.1 JH767162 EQC32639.1 BBXB01000228 GAY03090.1 KK583201 KDO30507.1 ANIZ01003263 ETI34925.1 ANIY01003905 ETP32927.1 ANJA01003445 ETO63688.1 KI688916 KI682423 KI695655 ETK75217.1 ETL81896.1 ETM35100.1 KI675716 ETL28642.1 ANIX01003713 ETP04802.1 KI669628 ETN01291.1 KI913996 ETV92918.1 ETV92919.1 KI913138 ETV76002.1 GL833120 EGB13141.1 CT867985 CAK56057.1 CCKQ01018262 CDW90215.1 LNFO01003828 KUF82153.1 CR548612 CAH03255.1 MBAD02001794 MBDO02000156 RLN51797.1 RLN61448.1 JH159152 EGZ22833.1
JTDY01000584 KOB76569.1 NNAY01000341 OXU29070.1 KQ981965 KYN31736.1 GL888243 EGI63954.1 KQ976542 KYM81060.1 ADTU01015125 LBMM01000169 KMR04642.1 KK107139 QOIP01000003 EZA57768.1 RLU24740.1 GECZ01012461 JAS57308.1 CM004474 OCT81553.1 PZQS01000001 PVD38475.1 CT868492 CAK83947.1 AMQN01011614 KB308980 ELT95875.1 KB202619 ESO89175.1 NEDP02005488 OWF40099.1 AFYH01115911 MLAK01000695 OHT07476.1 AAGJ04033126 DS113410 EAY06986.1 CT868062 CAK68702.1 JW871184 AFP03702.1 FR824259 CCA23946.1 CAIX01000156 CCI47185.1 CCI47183.1 GG662673 EAR97055.2 CCI47184.1 GL376595 GL983641 EGR32520.1 DS469512 EDO48733.1 JNBR01000127 OQR96839.1 GEBQ01002230 JAT37747.1 JH823224 EKC21326.1 JH767162 EQC32639.1 BBXB01000228 GAY03090.1 KK583201 KDO30507.1 ANIZ01003263 ETI34925.1 ANIY01003905 ETP32927.1 ANJA01003445 ETO63688.1 KI688916 KI682423 KI695655 ETK75217.1 ETL81896.1 ETM35100.1 KI675716 ETL28642.1 ANIX01003713 ETP04802.1 KI669628 ETN01291.1 KI913996 ETV92918.1 ETV92919.1 KI913138 ETV76002.1 GL833120 EGB13141.1 CT867985 CAK56057.1 CCKQ01018262 CDW90215.1 LNFO01003828 KUF82153.1 CR548612 CAH03255.1 MBAD02001794 MBDO02000156 RLN51797.1 RLN61448.1 JH159152 EGZ22833.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000037510
UP000002358
UP000215335
+ More
UP000078541 UP000007755 UP000078540 UP000005205 UP000036403 UP000053097 UP000279307 UP000192223 UP000085678 UP000186698 UP000245119 UP000000600 UP000014760 UP000030746 UP000242188 UP000008672 UP000007110 UP000001542 UP000053237 UP000087266 UP000009168 UP000008983 UP000001593 UP000243579 UP000005408 UP000030762 UP000054177 UP000030745 UP000018721 UP000018948 UP000028582 UP000053236 UP000054423 UP000054532 UP000053864 UP000018958 UP000018817 UP000024375 UP000019040 UP000052943 UP000277300 UP000284657 UP000002640
UP000078541 UP000007755 UP000078540 UP000005205 UP000036403 UP000053097 UP000279307 UP000192223 UP000085678 UP000186698 UP000245119 UP000000600 UP000014760 UP000030746 UP000242188 UP000008672 UP000007110 UP000001542 UP000053237 UP000087266 UP000009168 UP000008983 UP000001593 UP000243579 UP000005408 UP000030762 UP000054177 UP000030745 UP000018721 UP000018948 UP000028582 UP000053236 UP000054423 UP000054532 UP000053864 UP000018958 UP000018817 UP000024375 UP000019040 UP000052943 UP000277300 UP000284657 UP000002640
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
A0A3S2M9Z5
A0A194QQV1
A0A194Q872
A0A0L7LMI7
K7JNK0
A0A232FEX3
+ More
A0A195EUA2 F4WP23 A0A195BA04 A0A158NGL2 A0A0J7L9R5 A0A026WP81 A0A1W4X333 A0A1S3I933 A0A1B6G4E3 A0A1L8GCI2 A0A2T7PYH0 A0DLN0 R7TY40 V4A2K7 A0A210PUC3 H3AGS5 A0A1J4KCQ1 W4YHM2 A2EK02 A0CD35 V9KZ61 F0WRI3 A0A024GKW5 A0A024GK81 A0A1S3NFL6 Q23JY8 A0A024GKC3 K3X9H1 G0QQI5 A7RIL3 A0A1V9ZFR5 A0A1B6MP67 K1QI52 T0Q478 A0A2D4CBH1 A0A067CUN1 V9E791 W2YDS0 A0A080ZAM7 W2K9I3 W2I5E0 W2W2M1 W2PL20 A0A024TFX1 A0A024TG12 W4G8H5 F0XVH6 A0BBZ0 A0A078B6P6 A0A0W8CDI1 Q6BGG5 A0A3F2RP10 G4YR99
A0A195EUA2 F4WP23 A0A195BA04 A0A158NGL2 A0A0J7L9R5 A0A026WP81 A0A1W4X333 A0A1S3I933 A0A1B6G4E3 A0A1L8GCI2 A0A2T7PYH0 A0DLN0 R7TY40 V4A2K7 A0A210PUC3 H3AGS5 A0A1J4KCQ1 W4YHM2 A2EK02 A0CD35 V9KZ61 F0WRI3 A0A024GKW5 A0A024GK81 A0A1S3NFL6 Q23JY8 A0A024GKC3 K3X9H1 G0QQI5 A7RIL3 A0A1V9ZFR5 A0A1B6MP67 K1QI52 T0Q478 A0A2D4CBH1 A0A067CUN1 V9E791 W2YDS0 A0A080ZAM7 W2K9I3 W2I5E0 W2W2M1 W2PL20 A0A024TFX1 A0A024TG12 W4G8H5 F0XVH6 A0BBZ0 A0A078B6P6 A0A0W8CDI1 Q6BGG5 A0A3F2RP10 G4YR99
PDB
1P22
E-value=0.000130985,
Score=108
Ontologies
GO
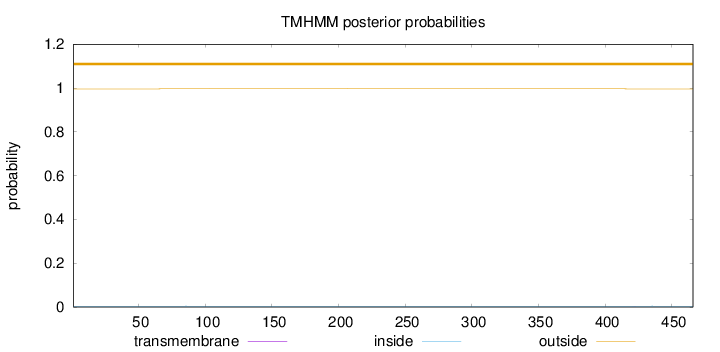
Topology
Length:
466
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00631000000000001
Exp number, first 60 AAs:
0.00039
Total prob of N-in:
0.00382
outside
1 - 466
Population Genetic Test Statistics
Pi
216.88839
Theta
169.83059
Tajima's D
1.055637
CLR
0.313974
CSRT
0.677666116694165
Interpretation
Uncertain
