Gene
KWMTBOMO05839
Pre Gene Modal
BGIBMGA001404
Annotation
PREDICTED:_RNA-directed_DNA_polymerase_from_mobile_element_jockey-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.887
Sequence
CDS
ATGTATAAACTACTTGAGCGAATCACACTTAACCGCATTGCACCAACTATAGAAAAACATATTCCTCAAGAGCAAGCTGGATTCCGTCCAGGACGCAGCTGCAGCGATCAAGTGTTAGCACTGACCACACTCTTGGAGAGGGGATTTAATGAAAACCTCAAGTCGTCAACTGTATTCGTGGACCTAACCGCTGCTTACGACACAGTATGGCGCCAGGGACTGCTGTACAAACTACTCAAGCTGATCCCCTGTATAAAGATCTACAAAGTTATTGAGGACGCTTTTACCAACAGACCATTCCGTGTCTATCTGGGAGACAAGTCTAGCTTATCCAGGGTCCTGAACAACGGTCTCCCGCAGGGCTCAGTCCTCGCACCAACACTCTTCAACGTGTACACCCACGATGTGCCAGAGATGCTCTCCAGGAAATTTCCGTACGCCGACGATCTAGCTCTAGTCGCACAAGGCCGCAGTATAGAGACAACGGAATGTCAGCTTGAAAACGACATGGCCACGCTTGATGACTACTTCATTCGATGGCGCTTGTGTCCAAACGCTTCCAAGACGGAAGTCTGCTGTTTCCACCTAAATAACCGACAGGCTAACAAGAAACCCAACGTCACCTTTAGAGGTTCCACTCTCCCCTTTAACCCCTATCCCAAATATCTGAGTGTCACCCTAGACCTGGTTTCAGAGAAAAACGCCCAATCTGGTGCATGCTTAACCACCTGA
Protein
MYKLLERITLNRIAPTIEKHIPQEQAGFRPGRSCSDQVLALTTLLERGFNENLKSSTVFVDLTAAYDTVWRQGLLYKLLKLIPCIKIYKVIEDAFTNRPFRVYLGDKSSLSRVLNNGLPQGSVLAPTLFNVYTHDVPEMLSRKFPYADDLALVAQGRSIETTECQLENDMATLDDYFIRWRLCPNASKTEVCCFHLNNRQANKKPNVTFRGSTLPFNPYPKYLSVTLDLVSEKNAQSGACLTT
Summary
Uniprot
A0A2S2R704
H9ISG9
A0A2S2R976
A0A2S2R253
D7GXJ1
A0A2W5AJS5
+ More
A0A2S2PC59 A0A2H8TVX2 X1WVV2 A0A1B6EN51 A0A2S2R0G0 J9L403 J9LNS0 A0A2S2QUG9 J9M1G4 J9KEZ9 A0A1B6KBV5 A0A1B6MIJ0 J9M8I0 A0A1B0D7V9 J9KK83 A0A0A9XVR4 J9L8J9 X1WPT5 A0A2S2Q452 J9KCY3 A0A2A4JQG8 A0A2S2QXK4 A0A2S2QZD0 J9L783 D6WE86 A0A2S2PKC8 X1WLF6 J9KT55 A0A2S2P4T9 W4YNK6 A0A2S2NDH1 J9M4Z0 W4XZG1 J9L8V9 A0A2W1BYF3 A0A3Q2YYG1 A0A3Q3E6Q2 J9L1U8 J9LFU6 W4XC16 A0A3B4VIU4 A0A2T7PMN6 J9LDH1 A0A2S2PR14 A0A0J7MT32 A0A2W1BC05 X1X928 A0A3S1ASJ4 A0A0A9Y6N1 X1WUG2 A0A023EW17 A0A2S2Q823 A0A0A9YHZ1 A0A1Y1NA23 A0A1Q3G5J4 A0A1Q3G5F7 J9KCZ4 J9KKL3 A0A2A4IUM2 V9GZF1 Q07995 A0A2S2N814 J9KIX1 A0A146KRW8 A0A0A9YSX4 X1WNI2 X1WXU3 A0A3Q2YP85 A0A2S2QV02 X1WWC4 J9LC21 A0A0A9YHY8 V5GNF3
A0A2S2PC59 A0A2H8TVX2 X1WVV2 A0A1B6EN51 A0A2S2R0G0 J9L403 J9LNS0 A0A2S2QUG9 J9M1G4 J9KEZ9 A0A1B6KBV5 A0A1B6MIJ0 J9M8I0 A0A1B0D7V9 J9KK83 A0A0A9XVR4 J9L8J9 X1WPT5 A0A2S2Q452 J9KCY3 A0A2A4JQG8 A0A2S2QXK4 A0A2S2QZD0 J9L783 D6WE86 A0A2S2PKC8 X1WLF6 J9KT55 A0A2S2P4T9 W4YNK6 A0A2S2NDH1 J9M4Z0 W4XZG1 J9L8V9 A0A2W1BYF3 A0A3Q2YYG1 A0A3Q3E6Q2 J9L1U8 J9LFU6 W4XC16 A0A3B4VIU4 A0A2T7PMN6 J9LDH1 A0A2S2PR14 A0A0J7MT32 A0A2W1BC05 X1X928 A0A3S1ASJ4 A0A0A9Y6N1 X1WUG2 A0A023EW17 A0A2S2Q823 A0A0A9YHZ1 A0A1Y1NA23 A0A1Q3G5J4 A0A1Q3G5F7 J9KCZ4 J9KKL3 A0A2A4IUM2 V9GZF1 Q07995 A0A2S2N814 J9KIX1 A0A146KRW8 A0A0A9YSX4 X1WNI2 X1WXU3 A0A3Q2YP85 A0A2S2QV02 X1WWC4 J9LC21 A0A0A9YHY8 V5GNF3
Pubmed
EMBL
GGMS01015939
MBY85142.1
BABH01040680
GGMS01017393
MBY86596.1
GGMS01014914
+ More
MBY84117.1 GG694201 EFA13393.1 QFNT01000080 PZO93396.1 GGMR01011027 GGMR01014159 MBY23646.1 MBY26778.1 GFXV01006156 MBW17961.1 ABLF02041704 GECZ01030418 JAS39351.1 GGMS01014285 MBY83488.1 ABLF02018295 ABLF02048731 ABLF02008383 ABLF02008387 ABLF02008388 ABLF02008389 ABLF02042376 ABLF02043653 GGMS01011579 MBY80782.1 ABLF02008152 ABLF02025872 ABLF02015153 ABLF02015154 GEBQ01031055 JAT08922.1 GEBQ01004298 JAT35679.1 ABLF02029964 AJVK01031149 ABLF02021396 GBHO01020646 JAG22958.1 ABLF02032924 ABLF02013865 ABLF02013866 ABLF02059691 GGMS01002779 MBY71982.1 ABLF02009760 NWSH01000896 PCG73650.1 GGMS01013298 MBY82501.1 GGMS01013279 MBY82482.1 ABLF02031655 KQ971325 EEZ99467.2 GGMR01017280 MBY29899.1 ABLF02030901 ABLF02059692 ABLF02033421 GGMR01011854 MBY24473.1 AAGJ04013553 GGMR01002645 MBY15264.1 ABLF02007187 ABLF02020407 ABLF02051791 ABLF02064862 AAGJ04063984 ABLF02014865 ABLF02025557 ABLF02067079 KZ149907 PZC78267.1 ABLF02007188 ABLF02035831 ABLF02035833 ABLF02057398 AAGJ04076310 PZQS01000003 PVD34672.1 ABLF02035448 GGMR01019198 MBY31817.1 LBMM01018951 KMQ83660.1 KZ150236 PZC71961.1 ABLF02066620 RQTK01001306 RUS70916.1 GBHO01016303 JAG27301.1 ABLF02016613 GAPW01000276 JAC13322.1 GGMS01004685 MBY73888.1 GBHO01014486 GBHO01014484 GBHO01012635 JAG29118.1 JAG29120.1 JAG30969.1 GEZM01008457 JAV94772.1 GFDL01000006 JAV35039.1 GFDL01000004 JAV35041.1 ABLF02008463 ABLF02029306 ABLF02029314 ABLF02060138 NWSH01007753 PCG62823.1 M95171 AAA29354.1 S59870 AAB26437.2 GGMR01000676 MBY13295.1 ABLF02030646 GDHC01020244 JAP98384.1 GBHO01008295 JAG35309.1 ABLF02008153 ABLF02008462 ABLF02008464 ABLF02008933 GGMS01012362 MBY81565.1 ABLF02003961 ABLF02059872 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 GBHO01014489 GBHO01012641 JAG29115.1 JAG30963.1 GALX01005359 JAB63107.1
MBY84117.1 GG694201 EFA13393.1 QFNT01000080 PZO93396.1 GGMR01011027 GGMR01014159 MBY23646.1 MBY26778.1 GFXV01006156 MBW17961.1 ABLF02041704 GECZ01030418 JAS39351.1 GGMS01014285 MBY83488.1 ABLF02018295 ABLF02048731 ABLF02008383 ABLF02008387 ABLF02008388 ABLF02008389 ABLF02042376 ABLF02043653 GGMS01011579 MBY80782.1 ABLF02008152 ABLF02025872 ABLF02015153 ABLF02015154 GEBQ01031055 JAT08922.1 GEBQ01004298 JAT35679.1 ABLF02029964 AJVK01031149 ABLF02021396 GBHO01020646 JAG22958.1 ABLF02032924 ABLF02013865 ABLF02013866 ABLF02059691 GGMS01002779 MBY71982.1 ABLF02009760 NWSH01000896 PCG73650.1 GGMS01013298 MBY82501.1 GGMS01013279 MBY82482.1 ABLF02031655 KQ971325 EEZ99467.2 GGMR01017280 MBY29899.1 ABLF02030901 ABLF02059692 ABLF02033421 GGMR01011854 MBY24473.1 AAGJ04013553 GGMR01002645 MBY15264.1 ABLF02007187 ABLF02020407 ABLF02051791 ABLF02064862 AAGJ04063984 ABLF02014865 ABLF02025557 ABLF02067079 KZ149907 PZC78267.1 ABLF02007188 ABLF02035831 ABLF02035833 ABLF02057398 AAGJ04076310 PZQS01000003 PVD34672.1 ABLF02035448 GGMR01019198 MBY31817.1 LBMM01018951 KMQ83660.1 KZ150236 PZC71961.1 ABLF02066620 RQTK01001306 RUS70916.1 GBHO01016303 JAG27301.1 ABLF02016613 GAPW01000276 JAC13322.1 GGMS01004685 MBY73888.1 GBHO01014486 GBHO01014484 GBHO01012635 JAG29118.1 JAG29120.1 JAG30969.1 GEZM01008457 JAV94772.1 GFDL01000006 JAV35039.1 GFDL01000004 JAV35041.1 ABLF02008463 ABLF02029306 ABLF02029314 ABLF02060138 NWSH01007753 PCG62823.1 M95171 AAA29354.1 S59870 AAB26437.2 GGMR01000676 MBY13295.1 ABLF02030646 GDHC01020244 JAP98384.1 GBHO01008295 JAG35309.1 ABLF02008153 ABLF02008462 ABLF02008464 ABLF02008933 GGMS01012362 MBY81565.1 ABLF02003961 ABLF02059872 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 GBHO01014489 GBHO01012641 JAG29115.1 JAG30963.1 GALX01005359 JAB63107.1
Proteomes
PRIDE
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR011989 ARM-like
IPR000225 Armadillo
IPR028435 Plakophilin/d_Catenin
IPR028446 JAC-1
IPR016024 ARM-type_fold
IPR003656 Znf_BED
IPR008906 HATC_C_dom
IPR036236 Znf_C2H2_sf
IPR011010 DNA_brk_join_enz
IPR013762 Integrase-like_cat_sf
IPR026630 EPM2A-int_1
IPR012337 RNaseH-like_sf
IPR036397 RNaseH_sf
IPR002156 RNaseH_domain
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR011989 ARM-like
IPR000225 Armadillo
IPR028435 Plakophilin/d_Catenin
IPR028446 JAC-1
IPR016024 ARM-type_fold
IPR003656 Znf_BED
IPR008906 HATC_C_dom
IPR036236 Znf_C2H2_sf
IPR011010 DNA_brk_join_enz
IPR013762 Integrase-like_cat_sf
IPR026630 EPM2A-int_1
IPR012337 RNaseH-like_sf
IPR036397 RNaseH_sf
IPR002156 RNaseH_domain
SUPFAM
Gene 3D
ProteinModelPortal
A0A2S2R704
H9ISG9
A0A2S2R976
A0A2S2R253
D7GXJ1
A0A2W5AJS5
+ More
A0A2S2PC59 A0A2H8TVX2 X1WVV2 A0A1B6EN51 A0A2S2R0G0 J9L403 J9LNS0 A0A2S2QUG9 J9M1G4 J9KEZ9 A0A1B6KBV5 A0A1B6MIJ0 J9M8I0 A0A1B0D7V9 J9KK83 A0A0A9XVR4 J9L8J9 X1WPT5 A0A2S2Q452 J9KCY3 A0A2A4JQG8 A0A2S2QXK4 A0A2S2QZD0 J9L783 D6WE86 A0A2S2PKC8 X1WLF6 J9KT55 A0A2S2P4T9 W4YNK6 A0A2S2NDH1 J9M4Z0 W4XZG1 J9L8V9 A0A2W1BYF3 A0A3Q2YYG1 A0A3Q3E6Q2 J9L1U8 J9LFU6 W4XC16 A0A3B4VIU4 A0A2T7PMN6 J9LDH1 A0A2S2PR14 A0A0J7MT32 A0A2W1BC05 X1X928 A0A3S1ASJ4 A0A0A9Y6N1 X1WUG2 A0A023EW17 A0A2S2Q823 A0A0A9YHZ1 A0A1Y1NA23 A0A1Q3G5J4 A0A1Q3G5F7 J9KCZ4 J9KKL3 A0A2A4IUM2 V9GZF1 Q07995 A0A2S2N814 J9KIX1 A0A146KRW8 A0A0A9YSX4 X1WNI2 X1WXU3 A0A3Q2YP85 A0A2S2QV02 X1WWC4 J9LC21 A0A0A9YHY8 V5GNF3
A0A2S2PC59 A0A2H8TVX2 X1WVV2 A0A1B6EN51 A0A2S2R0G0 J9L403 J9LNS0 A0A2S2QUG9 J9M1G4 J9KEZ9 A0A1B6KBV5 A0A1B6MIJ0 J9M8I0 A0A1B0D7V9 J9KK83 A0A0A9XVR4 J9L8J9 X1WPT5 A0A2S2Q452 J9KCY3 A0A2A4JQG8 A0A2S2QXK4 A0A2S2QZD0 J9L783 D6WE86 A0A2S2PKC8 X1WLF6 J9KT55 A0A2S2P4T9 W4YNK6 A0A2S2NDH1 J9M4Z0 W4XZG1 J9L8V9 A0A2W1BYF3 A0A3Q2YYG1 A0A3Q3E6Q2 J9L1U8 J9LFU6 W4XC16 A0A3B4VIU4 A0A2T7PMN6 J9LDH1 A0A2S2PR14 A0A0J7MT32 A0A2W1BC05 X1X928 A0A3S1ASJ4 A0A0A9Y6N1 X1WUG2 A0A023EW17 A0A2S2Q823 A0A0A9YHZ1 A0A1Y1NA23 A0A1Q3G5J4 A0A1Q3G5F7 J9KCZ4 J9KKL3 A0A2A4IUM2 V9GZF1 Q07995 A0A2S2N814 J9KIX1 A0A146KRW8 A0A0A9YSX4 X1WNI2 X1WXU3 A0A3Q2YP85 A0A2S2QV02 X1WWC4 J9LC21 A0A0A9YHY8 V5GNF3
PDB
6AR3
E-value=6.49755e-05,
Score=108
Ontologies
KEGG
GO
PANTHER
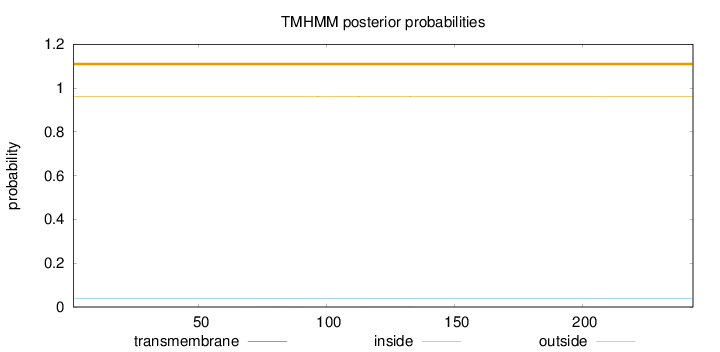
Topology
Length:
243
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01054
Exp number, first 60 AAs:
0.0005
Total prob of N-in:
0.03909
outside
1 - 243
Population Genetic Test Statistics
Pi
132.879621
Theta
54.374501
Tajima's D
1.11363
CLR
11.262216
CSRT
0.694115294235288
Interpretation
Uncertain
