Gene
KWMTBOMO05834 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006905
Annotation
alpha-L-fucoside_fucohydrolase_[Spodoptera_litura]
Location in the cell
Lysosomal Reliability : 1.44
Sequence
CDS
ATGAAGTCATTTTTCTTGGTATTCTTATTCGGATTCGTGAACGGCGCCATACACAATCGTCGTGATCATAGTTCGCATAATAAAATACTAAATAAGCGGTATTCTCCTGATTGGAACGATTTGGATACACGACCGTTGCCAGAGTGGTATGATAAAGCAAAAATAGGCATTTTCCTACATTGGGGCGTCTATTCTGTGCCCGGTTTTGGATCCGAATGGTTTTGGAGCAATTGGAAAGGTGGTAACCGACAATGCATCGATTTCATGACGAAAAATTATCCTCCAGGATTCACCTATCAGGAATTCGCACCCGCATTCAAAGCCGAGTTTTTCGATCCGGACAAATGGGCGGATCTTTTCGTAAAAGCCGGAGCAAAATACGTAGTGCTAACAAGTAAACATCATGAGGGCTTCACTTTGTTCCCGTCAAAACGTTCGTTCAGTTGGAATTCCGTCGAAGTGGGCCCAAAACGCGATATCGTCGGGGACCTGGCTAAGGCGGTGCGAAACACAAGCTTGAAATTCGGCGTTTATCATTCGCTTTACGAGTGGTTCAACCCTATTTACGAAGAGGACAAGAGGAAGGCCTTCAAAACTCAGACGTACGTTGACACGAAACTGTGGCCGGACATAAAACAGATCGTTCACGATTACAAGCCCTCCGTTATATGGTCTGACGGTGATTGGGAAGCGAACGACACCTACTGGCGGTCCACGGAACTGCTCGCCTGGTTGTACAACGACAGCCCTGTCAAAGACGAAGTAGTCGTCAACGATCGTTGGGGCATCGGCATACCAGGAAAGCACGGCGATTTCTACAATCATCAAGACAGATTTAACCCTGGAGTCCTTTTGCACCATAAGTGGGAAAACGCGTTCACTGTGGACTCCGGATCTTGGGGATACAGGCGGAATATGCAAATACGCGAAATATTGACGATTGAAGAATTGCTCCAACAAATCGTAACAACAGTTAGCTGCGGGGGAAACGCGCTCATAAATGTCGGTCCAACTAAAGAAGGTACGATCGCCCCGATATTTGAGGAACGACTTTTGGCGCTAGGCGACTGGCTCCAAATAAACGGCGAGGCCATATACAGCTCCTCGCCGTGGGAGTATCAAAACGACACTCTAAATGGCAAAGTTTGGTACACTTGTTCGATTAATAGAAGCAATAGTATGAAACGTACATACCGCAGTGGACAAGTGACTGCAATCTACGCTATTTTACTCGAATGGCCTAGCGACAACAGACTATCCGTTCGAAGTATCACAAACTCTTTGCGCAGTAGGTACAACCAACTTGAGATGTTAGGCAACGAAGGAACATACTTAAATTGGAAGATTACGAATGGGGACGTTGAAATTGAGCTTCCAGATAGAGCAACAGTAAAATCAAATCACGCTTGGGTCCTCAAACTCGCGACCACATAA
Protein
MKSFFLVFLFGFVNGAIHNRRDHSSHNKILNKRYSPDWNDLDTRPLPEWYDKAKIGIFLHWGVYSVPGFGSEWFWSNWKGGNRQCIDFMTKNYPPGFTYQEFAPAFKAEFFDPDKWADLFVKAGAKYVVLTSKHHEGFTLFPSKRSFSWNSVEVGPKRDIVGDLAKAVRNTSLKFGVYHSLYEWFNPIYEEDKRKAFKTQTYVDTKLWPDIKQIVHDYKPSVIWSDGDWEANDTYWRSTELLAWLYNDSPVKDEVVVNDRWGIGIPGKHGDFYNHQDRFNPGVLLHHKWENAFTVDSGSWGYRRNMQIREILTIEELLQQIVTTVSCGGNALINVGPTKEGTIAPIFEERLLALGDWLQINGEAIYSSSPWEYQNDTLNGKVWYTCSINRSNSMKRTYRSGQVTAIYAILLEWPSDNRLSVRSITNSLRSRYNQLEMLGNEGTYLNWKITNGDVEIELPDRATVKSNHAWVLKLATT
Summary
Similarity
Belongs to the glycosyl hydrolase 29 family.
Uniprot
H9JBL0
A0A2A4ISS5
S5G3E0
A0A2H1WT93
A0A194QQT1
I4DJT3
+ More
A0A194QE59 A0A3S2PLH2 A0A139WLC8 A0A1Y1KFM4 A0A194Q9R1 J3JXT8 V5GJB8 A0A067RQD7 V5HEY6 V5H5I0 V5II28 A0A0M8ZVM1 A0A0P5N9Z1 A0A182XYI1 W5JS94 A0A0P5EPC0 E9H8P9 A0A3M7R5S5 D6WGK5 A0A0P5VYW7 A0A0L7LJS9 A0A182T8J7 E2B4G2 A0A182FG61 A0A2M4BMU3 A0A182MTE9 A0A182WJM1 A0A2J7R465 A0A1B0CDN8 A0A310S8R9 A0A182RTY3 A0A139WKN6 A0A182G2D9 A0A2M4BM62 A0A0U4CIY7 N6UEU1 A0A2M3ZEZ4 A0A0P5L001 U4USR7 E9GY15 A0A182NKP0 B7QBK7 E0W0P0 A0A0P5BDH2 A0A026WMB6 A0A0P5LC60 A0A0N8E7Q6 A0A0P6CXM5 A0A2M3ZF06 A0A2M4CMG4 A0A1E1XA71 A0A0N8E275 A0A158NQ51 A0A182K058 A0A182KQU1 Q7QJ50 A0A182P218 A0A182I2V9 Q16S75 A0A182WZ47 F4WMS0 A0A182J2B0 A0A182UNZ3 A0A224YSV1 A0A1L8DQQ4 A0A1L8DQV4 A0A087TY18 A0A154PJQ2 A0A1S4FRE8 A0A131YJQ4 A0A232EVD8 A0A182UBR0 V5HAZ2 A0A088ADU2 T1IV28 A0A182Q4I5 A0A2A3E214 V9IE27 A0A158NQ52 A0A0L8GA35 A0A2M3ZI85 K1QFV3 A0A0L7R9D7 A0A0P5TTK4 A0A162S5I2 A0A0P5JDS0 A0A0P5SHC3 K7J648 H2ZMP0 H2ZMP2 A0A0P5MXY7 B0W3T8 A0A0P4VTZ8 A0A0P5XZP7
A0A194QE59 A0A3S2PLH2 A0A139WLC8 A0A1Y1KFM4 A0A194Q9R1 J3JXT8 V5GJB8 A0A067RQD7 V5HEY6 V5H5I0 V5II28 A0A0M8ZVM1 A0A0P5N9Z1 A0A182XYI1 W5JS94 A0A0P5EPC0 E9H8P9 A0A3M7R5S5 D6WGK5 A0A0P5VYW7 A0A0L7LJS9 A0A182T8J7 E2B4G2 A0A182FG61 A0A2M4BMU3 A0A182MTE9 A0A182WJM1 A0A2J7R465 A0A1B0CDN8 A0A310S8R9 A0A182RTY3 A0A139WKN6 A0A182G2D9 A0A2M4BM62 A0A0U4CIY7 N6UEU1 A0A2M3ZEZ4 A0A0P5L001 U4USR7 E9GY15 A0A182NKP0 B7QBK7 E0W0P0 A0A0P5BDH2 A0A026WMB6 A0A0P5LC60 A0A0N8E7Q6 A0A0P6CXM5 A0A2M3ZF06 A0A2M4CMG4 A0A1E1XA71 A0A0N8E275 A0A158NQ51 A0A182K058 A0A182KQU1 Q7QJ50 A0A182P218 A0A182I2V9 Q16S75 A0A182WZ47 F4WMS0 A0A182J2B0 A0A182UNZ3 A0A224YSV1 A0A1L8DQQ4 A0A1L8DQV4 A0A087TY18 A0A154PJQ2 A0A1S4FRE8 A0A131YJQ4 A0A232EVD8 A0A182UBR0 V5HAZ2 A0A088ADU2 T1IV28 A0A182Q4I5 A0A2A3E214 V9IE27 A0A158NQ52 A0A0L8GA35 A0A2M3ZI85 K1QFV3 A0A0L7R9D7 A0A0P5TTK4 A0A162S5I2 A0A0P5JDS0 A0A0P5SHC3 K7J648 H2ZMP0 H2ZMP2 A0A0P5MXY7 B0W3T8 A0A0P4VTZ8 A0A0P5XZP7
Pubmed
19121390
26354079
22651552
18362917
19820115
28004739
+ More
22516182 24845553 25765539 25244985 20920257 23761445 21292972 30375419 26227816 20798317 26483478 26694822 23537049 20566863 24508170 30249741 28503490 21347285 20966253 12364791 14747013 17210077 17510324 21719571 28797301 26830274 28648823 22992520 20075255
22516182 24845553 25765539 25244985 20920257 23761445 21292972 30375419 26227816 20798317 26483478 26694822 23537049 20566863 24508170 30249741 28503490 21347285 20966253 12364791 14747013 17210077 17510324 21719571 28797301 26830274 28648823 22992520 20075255
EMBL
BABH01024524
NWSH01007210
PCG63037.1
KC832290
AGQ53950.1
ODYU01010888
+ More
SOQ56273.1 KQ461175 KPJ07863.1 AK401551 BAM18173.1 KQ459302 KPJ01751.1 RSAL01000003 RVE54833.1 KQ971321 KYB28627.1 GEZM01087890 JAV58415.1 KPJ01750.1 BT128059 AEE63020.1 GALX01004282 JAB64184.1 KK852529 KDR21959.1 GANP01012465 JAB72003.1 GANP01014931 JAB69537.1 GANP01003912 JAB80556.1 KQ435828 KOX71867.1 GDIQ01148777 JAL02949.1 ADMH02000578 ETN65790.1 GDIP01143602 LRGB01000872 JAJ79800.1 KZS15372.1 GL732605 EFX71905.1 REGN01004207 RNA18598.1 EFA00166.1 GDIP01107794 JAL95920.1 JTDY01000854 KOB75665.1 GL445556 EFN89423.1 GGFJ01005259 MBW54400.1 AXCM01001909 NEVH01007815 PNF35620.1 AJWK01008050 KQ769940 OAD52751.1 KYB28628.1 JXUM01039705 KQ561215 KXJ79327.1 GGFJ01005026 MBW54167.1 KU218694 ALX00074.1 APGK01037834 KB740949 ENN77167.1 GGFM01006331 MBW27082.1 GDIQ01184770 JAK66955.1 KB632347 ERL93145.1 GL732574 EFX75639.1 ABJB010297592 ABJB010583610 DS902098 EEC16229.1 DS235862 EEB19196.1 GDIP01186735 JAJ36667.1 KK107167 QOIP01000010 EZA56254.1 RLU17743.1 GDIQ01171858 JAK79867.1 GDIQ01053620 JAN41117.1 GDIQ01086160 JAN08577.1 GGFM01006332 MBW27083.1 GGFL01002326 MBW66504.1 GFAC01003035 JAT96153.1 GDIQ01069068 JAN25669.1 ADTU01022963 AAAB01008807 EAA04031.4 APCN01000188 CH477680 EAT37340.1 GL888218 EGI64547.1 GFPF01005918 MAA17064.1 GFDF01005302 JAV08782.1 GFDF01005303 JAV08781.1 KK117271 KFM70007.1 KQ434938 KZC12089.1 GEDV01009759 JAP78798.1 NNAY01002011 OXU22311.1 GANP01003913 JAB80555.1 JH431572 AXCN02000549 KZ288497 PBC25179.1 JR041212 AEY59383.1 KQ423006 KOF73758.1 GGFM01007482 MBW28233.1 JH815850 EKC35757.1 KQ414627 KOC67449.1 GDIP01139748 JAL63966.1 LRGB01000084 KZS21030.1 GDIQ01206863 JAK44862.1 GDIP01198345 GDIP01139747 JAL63967.1 GDIQ01149679 JAL02047.1 DS231833 EDS32234.1 GDRN01110869 JAI56851.1 GDIP01064963 JAM38752.1
SOQ56273.1 KQ461175 KPJ07863.1 AK401551 BAM18173.1 KQ459302 KPJ01751.1 RSAL01000003 RVE54833.1 KQ971321 KYB28627.1 GEZM01087890 JAV58415.1 KPJ01750.1 BT128059 AEE63020.1 GALX01004282 JAB64184.1 KK852529 KDR21959.1 GANP01012465 JAB72003.1 GANP01014931 JAB69537.1 GANP01003912 JAB80556.1 KQ435828 KOX71867.1 GDIQ01148777 JAL02949.1 ADMH02000578 ETN65790.1 GDIP01143602 LRGB01000872 JAJ79800.1 KZS15372.1 GL732605 EFX71905.1 REGN01004207 RNA18598.1 EFA00166.1 GDIP01107794 JAL95920.1 JTDY01000854 KOB75665.1 GL445556 EFN89423.1 GGFJ01005259 MBW54400.1 AXCM01001909 NEVH01007815 PNF35620.1 AJWK01008050 KQ769940 OAD52751.1 KYB28628.1 JXUM01039705 KQ561215 KXJ79327.1 GGFJ01005026 MBW54167.1 KU218694 ALX00074.1 APGK01037834 KB740949 ENN77167.1 GGFM01006331 MBW27082.1 GDIQ01184770 JAK66955.1 KB632347 ERL93145.1 GL732574 EFX75639.1 ABJB010297592 ABJB010583610 DS902098 EEC16229.1 DS235862 EEB19196.1 GDIP01186735 JAJ36667.1 KK107167 QOIP01000010 EZA56254.1 RLU17743.1 GDIQ01171858 JAK79867.1 GDIQ01053620 JAN41117.1 GDIQ01086160 JAN08577.1 GGFM01006332 MBW27083.1 GGFL01002326 MBW66504.1 GFAC01003035 JAT96153.1 GDIQ01069068 JAN25669.1 ADTU01022963 AAAB01008807 EAA04031.4 APCN01000188 CH477680 EAT37340.1 GL888218 EGI64547.1 GFPF01005918 MAA17064.1 GFDF01005302 JAV08782.1 GFDF01005303 JAV08781.1 KK117271 KFM70007.1 KQ434938 KZC12089.1 GEDV01009759 JAP78798.1 NNAY01002011 OXU22311.1 GANP01003913 JAB80555.1 JH431572 AXCN02000549 KZ288497 PBC25179.1 JR041212 AEY59383.1 KQ423006 KOF73758.1 GGFM01007482 MBW28233.1 JH815850 EKC35757.1 KQ414627 KOC67449.1 GDIP01139748 JAL63966.1 LRGB01000084 KZS21030.1 GDIQ01206863 JAK44862.1 GDIP01198345 GDIP01139747 JAL63967.1 GDIQ01149679 JAL02047.1 DS231833 EDS32234.1 GDRN01110869 JAI56851.1 GDIP01064963 JAM38752.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007266
+ More
UP000027135 UP000053105 UP000076408 UP000000673 UP000076858 UP000000305 UP000276133 UP000037510 UP000075901 UP000008237 UP000069272 UP000075883 UP000075920 UP000235965 UP000092461 UP000075900 UP000069940 UP000249989 UP000019118 UP000030742 UP000075884 UP000001555 UP000009046 UP000053097 UP000279307 UP000005205 UP000075881 UP000075882 UP000007062 UP000075885 UP000075840 UP000008820 UP000076407 UP000007755 UP000075880 UP000075903 UP000054359 UP000076502 UP000215335 UP000075902 UP000005203 UP000075886 UP000242457 UP000053454 UP000005408 UP000053825 UP000002358 UP000007875 UP000002320
UP000027135 UP000053105 UP000076408 UP000000673 UP000076858 UP000000305 UP000276133 UP000037510 UP000075901 UP000008237 UP000069272 UP000075883 UP000075920 UP000235965 UP000092461 UP000075900 UP000069940 UP000249989 UP000019118 UP000030742 UP000075884 UP000001555 UP000009046 UP000053097 UP000279307 UP000005205 UP000075881 UP000075882 UP000007062 UP000075885 UP000075840 UP000008820 UP000076407 UP000007755 UP000075880 UP000075903 UP000054359 UP000076502 UP000215335 UP000075902 UP000005203 UP000075886 UP000242457 UP000053454 UP000005408 UP000053825 UP000002358 UP000007875 UP000002320
PRIDE
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
H9JBL0
A0A2A4ISS5
S5G3E0
A0A2H1WT93
A0A194QQT1
I4DJT3
+ More
A0A194QE59 A0A3S2PLH2 A0A139WLC8 A0A1Y1KFM4 A0A194Q9R1 J3JXT8 V5GJB8 A0A067RQD7 V5HEY6 V5H5I0 V5II28 A0A0M8ZVM1 A0A0P5N9Z1 A0A182XYI1 W5JS94 A0A0P5EPC0 E9H8P9 A0A3M7R5S5 D6WGK5 A0A0P5VYW7 A0A0L7LJS9 A0A182T8J7 E2B4G2 A0A182FG61 A0A2M4BMU3 A0A182MTE9 A0A182WJM1 A0A2J7R465 A0A1B0CDN8 A0A310S8R9 A0A182RTY3 A0A139WKN6 A0A182G2D9 A0A2M4BM62 A0A0U4CIY7 N6UEU1 A0A2M3ZEZ4 A0A0P5L001 U4USR7 E9GY15 A0A182NKP0 B7QBK7 E0W0P0 A0A0P5BDH2 A0A026WMB6 A0A0P5LC60 A0A0N8E7Q6 A0A0P6CXM5 A0A2M3ZF06 A0A2M4CMG4 A0A1E1XA71 A0A0N8E275 A0A158NQ51 A0A182K058 A0A182KQU1 Q7QJ50 A0A182P218 A0A182I2V9 Q16S75 A0A182WZ47 F4WMS0 A0A182J2B0 A0A182UNZ3 A0A224YSV1 A0A1L8DQQ4 A0A1L8DQV4 A0A087TY18 A0A154PJQ2 A0A1S4FRE8 A0A131YJQ4 A0A232EVD8 A0A182UBR0 V5HAZ2 A0A088ADU2 T1IV28 A0A182Q4I5 A0A2A3E214 V9IE27 A0A158NQ52 A0A0L8GA35 A0A2M3ZI85 K1QFV3 A0A0L7R9D7 A0A0P5TTK4 A0A162S5I2 A0A0P5JDS0 A0A0P5SHC3 K7J648 H2ZMP0 H2ZMP2 A0A0P5MXY7 B0W3T8 A0A0P4VTZ8 A0A0P5XZP7
A0A194QE59 A0A3S2PLH2 A0A139WLC8 A0A1Y1KFM4 A0A194Q9R1 J3JXT8 V5GJB8 A0A067RQD7 V5HEY6 V5H5I0 V5II28 A0A0M8ZVM1 A0A0P5N9Z1 A0A182XYI1 W5JS94 A0A0P5EPC0 E9H8P9 A0A3M7R5S5 D6WGK5 A0A0P5VYW7 A0A0L7LJS9 A0A182T8J7 E2B4G2 A0A182FG61 A0A2M4BMU3 A0A182MTE9 A0A182WJM1 A0A2J7R465 A0A1B0CDN8 A0A310S8R9 A0A182RTY3 A0A139WKN6 A0A182G2D9 A0A2M4BM62 A0A0U4CIY7 N6UEU1 A0A2M3ZEZ4 A0A0P5L001 U4USR7 E9GY15 A0A182NKP0 B7QBK7 E0W0P0 A0A0P5BDH2 A0A026WMB6 A0A0P5LC60 A0A0N8E7Q6 A0A0P6CXM5 A0A2M3ZF06 A0A2M4CMG4 A0A1E1XA71 A0A0N8E275 A0A158NQ51 A0A182K058 A0A182KQU1 Q7QJ50 A0A182P218 A0A182I2V9 Q16S75 A0A182WZ47 F4WMS0 A0A182J2B0 A0A182UNZ3 A0A224YSV1 A0A1L8DQQ4 A0A1L8DQV4 A0A087TY18 A0A154PJQ2 A0A1S4FRE8 A0A131YJQ4 A0A232EVD8 A0A182UBR0 V5HAZ2 A0A088ADU2 T1IV28 A0A182Q4I5 A0A2A3E214 V9IE27 A0A158NQ52 A0A0L8GA35 A0A2M3ZI85 K1QFV3 A0A0L7R9D7 A0A0P5TTK4 A0A162S5I2 A0A0P5JDS0 A0A0P5SHC3 K7J648 H2ZMP0 H2ZMP2 A0A0P5MXY7 B0W3T8 A0A0P4VTZ8 A0A0P5XZP7
PDB
2ZXD
E-value=1.30568e-63,
Score=617
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.972223
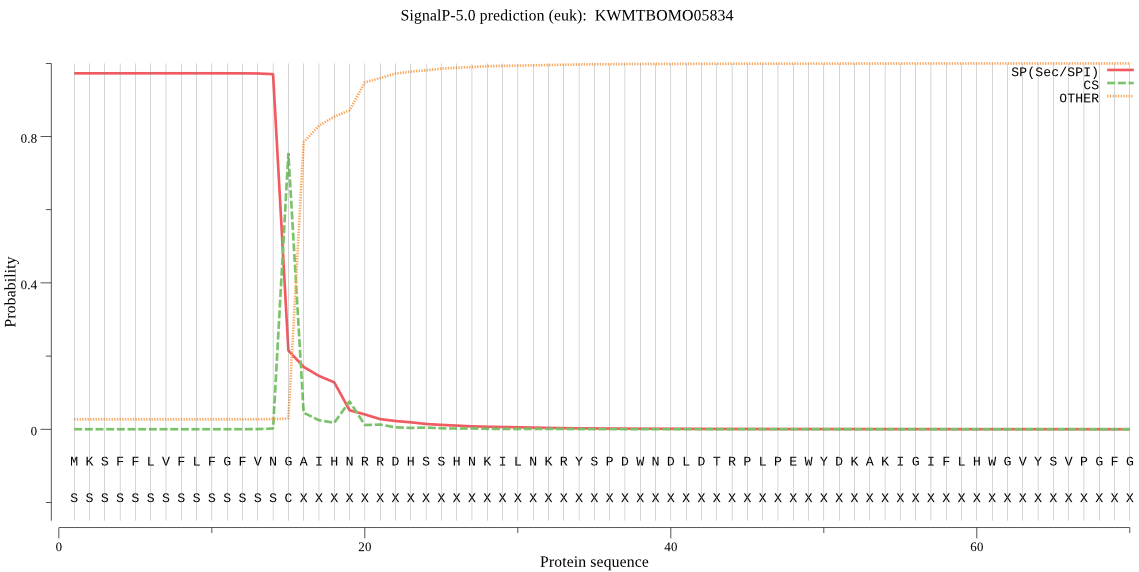
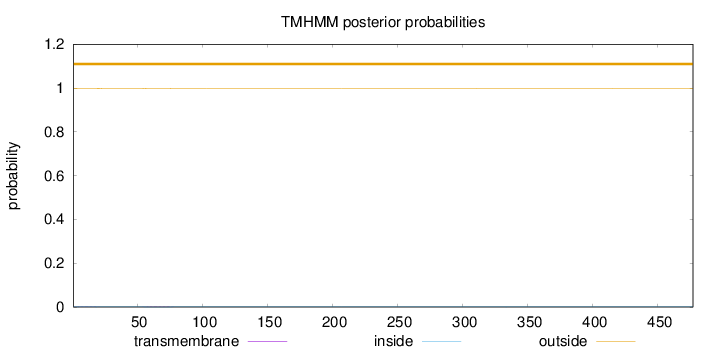
Length:
477
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05491
Exp number, first 60 AAs:
0.02885
Total prob of N-in:
0.00144
outside
1 - 477
Population Genetic Test Statistics
Pi
183.232949
Theta
205.746885
Tajima's D
-0.378315
CLR
0.461391
CSRT
0.269036548172591
Interpretation
Possibly Positive selection
