Pre Gene Modal
BGIBMGA006952
Annotation
PREDICTED:_TBC1_domain_family_member_9_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.523
Sequence
CDS
ATGTGGGTGAAACCTCAAGAAATATATTTGAAAACCGCACTATGGGATACGGAAGAAACATCTTTGTATTTTGTTTTACAATGTAGAAAAGGACATGGTAAATCAAAAGGGCTCTCCTCGCTTCTTGTTGGTACTTTAGACAGCGTCCGAGACACCAAACCTGCGCCCTACAGGATATTACATCAAACTCCAAACTCCGAAATCTATTATGTAATAGCAACTGCATTGACGGAACAACAGATCCGTCAAGATTGGCAGTGGCTTATTGACAATGTTTGTTCAACACTCCATTCCTTTGACACCGAGGAAGAAGTCACAGAATTTGTATGCTGCAAAATTAACTCTGTTATGGCAACTGAACAAGAAAATATAACTGAAGATGAAGACACAATAACTTACAAAAATGTTGATTATGAATTTCACCAAAGATTTAATATGCCCAAAGATGAAAAACTTGTGTGCTATTATTCATGCAGTTATTGGAAGGGAAAAATGCCACGTCAGGGATGGATGTATCTGTCTGTCCATTACATGTGTTTCTATTCATACATTATCGGTCGGAAAACAGCTGTTAAGATACGTTGGGCTGATGTGACAGAGCTCGCAAAAACACACAGCCTAATATTTCCAGACTCCATTAAAGTGGTGACCAGGGATGCTGAGCATTATTTCACTATGTTTTTGAAGAAGAATGAGACTTTTGCATTAATGCAGCAGCTCGCCAATATTGCTATGAAACAGCTTATAGATGATAAATCTGGAACCTTCAACATTGACAAAGATTTATTAACAAAACTGAGCAAAAATGTGCCAAAGAAAGCATCTTTTTTGAAACGTGACCTAGATGCTAGAAAGCAATCAAATCTGTACACACTAAGGTTCCGATTGCCTCAAACAGAAAAGCTTGATGGAACTGAAGCATGTACTCTCTGGACACCATACAATAAAAAACACAATTGGGGAAGACTATATTTGTCTCAGAATTTTATATGTTTTGACAGCAGGGTGCCAAATTTGGTACGACTGACTATCCCACTGCGCAACGTACATCAAGTGGAACGCGCTGACTCCGGCGGCACTGGAAGCTCAGGAGATTCTGGTAGCATCCTCATCACGACCGCCCACCACACCAGCTTCCTTTTCGGAAACATATCCGACAGGGATTTTCTTGTGCACAAAATCTCTGAGCTACTGGCAAAATTACCAAAGGAGAAGGCGATACGGGCAATGGCGAAACTCGAAAATGACAGTCACTGGACTCCGCAGCCCCCTTTGATGACGCTATTCAAGACTCATCAAACTTCACCGATTAAACAAATGCAACAGAAAAAGGAGAAAAGATGGGAAGACCACATGGCTGAAGTCGGTCGAGGGGTGAGCATGTACCAAACGACAGACGGTAGCGAATTAGTCGTCAGCGGCCTTCCTGAGTCTCTTCGTGGAGAACTATGGAGCGTGTTCTCGGGTTCTATACTGCAGAAGGCACAGAACAAAGGACTGTACGAAAGACTGGTTAATGAGGCTCTTTCATCGAAGAATCAGGCGAACGATGAAATAGAACGAGATTTGCACAGATCATTGCCGGAACATAGGGCATTTCAAAACGATATCGGAATATCGGCATTACGTCGGGTTCTATGTGCGTACGCTCGCAAGAATCCCACAATAGGATATTGTCAGGCGATGAACATCGTGGCGTCTGTACTACTCATATATTGTCCGGAAGAGCAGGCGTTCTGGTTGTTAGCAACAATATGCGAGACTTTGTTGCCAGATTACTACAATACCAGGGTAGTCGGAGCTCTGGTCGATCAAGGAGTCCTTGAAGAACTAACAGAAGCTCATCTGCCGGAATTACACGCGAAGCTAGACGAACTGGGAATGATGAAAATGATCAGTCTGTCTTGGTTCCTGACTCTGTTCATCAGCGTGATGCCGTACGAGTGCGCGGTCAACGTTATGGACTGTTTCTTTTATGATGGCGCTAAAGTCATATTCCAGGTAACGCTCACGATTCTCGACATCAATCACGAGAAGCTACTCAAATGCACCGAAGATGGACAGGCTATGCAGGTTTTAAACGAGTATCTAGAAGGCATTTACAACGAGGAAGCAATGGCGCTGTCCACAGAACCACGAACTGAGGCAAAAACAATAAAGATTCAAGATCTCATTTACCGTACTTACCGTTCGTACGGCAGCACCGTTACCAGCGAACATATAGAGCACTTGCGACTGAAACACCGCCTGCGGGTAGTCAAACAACTAGAGGACAGCCTGGAGAGGAATACATTGAGGGCTTTACAACAAGACAAGCTGCTGGAGCCTAATGAATTGCATGATCTCTTAAGCATAATACGCGAAGAGTTATTCTGGGCGAAGCGTGTACCTTGCGAAAGATTGGAAGCTCCTTACGAAGCTTACCGACTCGATTACACCCAATTCAAAAGTTTATTCACTGCTCTATCGCCTTGGGCCAAAGGGGATTACGCCGAAGCTATTACTGCTAGAATGTTTAAGTTAATGGACCACGACGACGACGGGTTCGTGAGCGCTCGCGGCCTGAGCGTGGCCATAGGGCTGAGCACCCGCGGCGACGCCCAGCGCCGGCTGCGCGCCCTGTACTGCGCGCACGCGCCGCCGCTGCTCACCCACCAGGACCTCAGGCCCACGCACTTCACGGAAACCGGGGAAGAGGTCGCCGCCGACGCCATCTCCTTCTTCGACGACATAGAAAATCCACCAACGCCAGAACCATTAGCGCTGTCAGGTCTGCAGCGATCGATAAGTGAAGTGTGCGGTGAAGGTAGCAACATGGATAAGTCCACTGACAGCACCACAAGTCAAACCGGCTCGCCATGGGAGGGCTACACGTCGGAGTCGGTCCGTGCGTGCGGACTAGCCGGACTGGCGGGGTCGTCGCCCCCGAGCCCGGGCCCCGCCCCGCCCCCTCCCGCGCCGCCGCTCCCCCGCGCGCACTTCCTGCAGCTCCTCGCCGCGCTGCACGACCTCACGCAGCACAGCGCGCCGCACTACGAAGCCGTCGTCTTGGCAGGCACCTTGCTGTTACAACTCGGCGAGTTAAACCACCGGAGGCCGGATCTGGAGCGAGAAATGTCCCAGGACAGTCTGTACCTGGCGGCCGCGGCGAAGCAGTCGTCCACTTCCACGATGGATCCAAACGGCAACACCCCAGACGAACAAGAAATTCGAAGGACGCAGTCTCAAGAGTCTGAAAGCGTTTGGTCGATAACGTTAGAACAATTCCTGGCCACAATGCTTCTGCAGCCACCCCTAGAAGAATACTTCAACGAGAAGATATCTCTGGTGCCGCAACTGGAAGCGTTGAAGCAAAGAGATCGGCTCCACTCCGTCTCGTAG
Protein
MWVKPQEIYLKTALWDTEETSLYFVLQCRKGHGKSKGLSSLLVGTLDSVRDTKPAPYRILHQTPNSEIYYVIATALTEQQIRQDWQWLIDNVCSTLHSFDTEEEVTEFVCCKINSVMATEQENITEDEDTITYKNVDYEFHQRFNMPKDEKLVCYYSCSYWKGKMPRQGWMYLSVHYMCFYSYIIGRKTAVKIRWADVTELAKTHSLIFPDSIKVVTRDAEHYFTMFLKKNETFALMQQLANIAMKQLIDDKSGTFNIDKDLLTKLSKNVPKKASFLKRDLDARKQSNLYTLRFRLPQTEKLDGTEACTLWTPYNKKHNWGRLYLSQNFICFDSRVPNLVRLTIPLRNVHQVERADSGGTGSSGDSGSILITTAHHTSFLFGNISDRDFLVHKISELLAKLPKEKAIRAMAKLENDSHWTPQPPLMTLFKTHQTSPIKQMQQKKEKRWEDHMAEVGRGVSMYQTTDGSELVVSGLPESLRGELWSVFSGSILQKAQNKGLYERLVNEALSSKNQANDEIERDLHRSLPEHRAFQNDIGISALRRVLCAYARKNPTIGYCQAMNIVASVLLIYCPEEQAFWLLATICETLLPDYYNTRVVGALVDQGVLEELTEAHLPELHAKLDELGMMKMISLSWFLTLFISVMPYECAVNVMDCFFYDGAKVIFQVTLTILDINHEKLLKCTEDGQAMQVLNEYLEGIYNEEAMALSTEPRTEAKTIKIQDLIYRTYRSYGSTVTSEHIEHLRLKHRLRVVKQLEDSLERNTLRALQQDKLLEPNELHDLLSIIREELFWAKRVPCERLEAPYEAYRLDYTQFKSLFTALSPWAKGDYAEAITARMFKLMDHDDDGFVSARGLSVAIGLSTRGDAQRRLRALYCAHAPPLLTHQDLRPTHFTETGEEVAADAISFFDDIENPPTPEPLALSGLQRSISEVCGEGSNMDKSTDSTTSQTGSPWEGYTSESVRACGLAGLAGSSPPSPGPAPPPPAPPLPRAHFLQLLAALHDLTQHSAPHYEAVVLAGTLLLQLGELNHRRPDLEREMSQDSLYLAAAAKQSSTSTMDPNGNTPDEQEIRRTQSQESESVWSITLEQFLATMLLQPPLEEYFNEKISLVPQLEALKQRDRLHSVS
Summary
Uniprot
H9JBQ7
A0A194Q888
A0A194QQU1
A0A1E1W1V0
A0A212EI24
A0A2A4IX64
+ More
A0A2A4IUJ7 A0A2H1WV40 A0A158NCG7 A0A088AVK6 A0A2A4IYK2 A0A2J7PYF0 E2C358 A0A0C9RRM3 A0A0N0U388 A0A0L7RJ63 A0A2J7PYF6 A0A154PQ11 K7IPM7 A0A195BXB1 A0A1Y1KBA7 A0A232EMQ4 E2AD73 A0A3L8DI37 A0A151WNC2 A0A1B6E0Q9 A0A1B6C1U8 A0A2A3EPJ6 E9IM53 A0A151IBY4 A0A023F161 A0A069DX64 A0A195F8U1 T1HS16 T1JBL6 A0A1E1XPN5 A0A131XGW9 A0A224Z875 A0A2R5LEK8 L7MKW6 A0A131XYW0 E0VCZ6 L7ML78 A0A147BT99 A0A226EKI2 A0A1S3JSL2 R7TPZ1 A0A1S3JSN9 U4U0V3 J9K185 A0A2H8TX45 A0A1E1XHV0 C3YSA6 A0A2C9K5E8 A0A2S2R7Z9 A0A1B6HYF8 A0A210QPU5 A0A0B6ZTR9 A0A1D2MTT0 V4C7V3 A0A3B3T1T3 A0A3B3T105 A0A3B3T1Y9 W5N0E3 A0A026WUX3 A0A1L8DMK7 A0A1L8DMP1 A0A0F7Z5H4 A0A3B3T377 A0A3Q2XHJ3 A0A3B3T1E2 A0A3B3T320 A0A1W4YMM1 A0A2T7PJ41 A0A0P7V1N1 A0A1A7XZX8 A0A3B4EMN8 E7F7E8 A0A1W4YWN3 A0A1A7WB39 A0A286Y906 A0A182GID6 A0A182QVV2 A0A2D0SEL7 A0A182J9L3 A0A3B4ZMZ6 A0A2D0SDW3 A0A3B4DBT0 A0A3B3T167 G3PYY0 F1S448 A0A287B5J0 A0A132AE57 A0A3P8X003 A0A1A8FHX3 A0A1Q3FXT6 A0A1W3JAA4 A0A1A8N900
A0A2A4IUJ7 A0A2H1WV40 A0A158NCG7 A0A088AVK6 A0A2A4IYK2 A0A2J7PYF0 E2C358 A0A0C9RRM3 A0A0N0U388 A0A0L7RJ63 A0A2J7PYF6 A0A154PQ11 K7IPM7 A0A195BXB1 A0A1Y1KBA7 A0A232EMQ4 E2AD73 A0A3L8DI37 A0A151WNC2 A0A1B6E0Q9 A0A1B6C1U8 A0A2A3EPJ6 E9IM53 A0A151IBY4 A0A023F161 A0A069DX64 A0A195F8U1 T1HS16 T1JBL6 A0A1E1XPN5 A0A131XGW9 A0A224Z875 A0A2R5LEK8 L7MKW6 A0A131XYW0 E0VCZ6 L7ML78 A0A147BT99 A0A226EKI2 A0A1S3JSL2 R7TPZ1 A0A1S3JSN9 U4U0V3 J9K185 A0A2H8TX45 A0A1E1XHV0 C3YSA6 A0A2C9K5E8 A0A2S2R7Z9 A0A1B6HYF8 A0A210QPU5 A0A0B6ZTR9 A0A1D2MTT0 V4C7V3 A0A3B3T1T3 A0A3B3T105 A0A3B3T1Y9 W5N0E3 A0A026WUX3 A0A1L8DMK7 A0A1L8DMP1 A0A0F7Z5H4 A0A3B3T377 A0A3Q2XHJ3 A0A3B3T1E2 A0A3B3T320 A0A1W4YMM1 A0A2T7PJ41 A0A0P7V1N1 A0A1A7XZX8 A0A3B4EMN8 E7F7E8 A0A1W4YWN3 A0A1A7WB39 A0A286Y906 A0A182GID6 A0A182QVV2 A0A2D0SEL7 A0A182J9L3 A0A3B4ZMZ6 A0A2D0SDW3 A0A3B4DBT0 A0A3B3T167 G3PYY0 F1S448 A0A287B5J0 A0A132AE57 A0A3P8X003 A0A1A8FHX3 A0A1Q3FXT6 A0A1W3JAA4 A0A1A8N900
Pubmed
EMBL
BABH01024523
BABH01024524
KQ459302
KPJ01752.1
KQ461175
KPJ07862.1
+ More
GDQN01010089 JAT80965.1 AGBW02014723 OWR41127.1 NWSH01005395 PCG64249.1 NWSH01007754 PCG62820.1 ODYU01010888 SOQ56274.1 ADTU01011884 PCG64250.1 NEVH01020353 PNF21369.1 GL452292 EFN77590.1 GBYB01010011 GBYB01010013 JAG79778.1 JAG79780.1 KQ435922 KOX68432.1 KQ414582 KOC70864.1 PNF21368.1 KQ434998 KZC13338.1 KQ976396 KYM92930.1 GEZM01087290 JAV58793.1 NNAY01003342 OXU19592.1 GL438684 EFN68621.1 QOIP01000008 RLU19823.1 KQ982934 KYQ49185.1 GEDC01011289 GEDC01005799 JAS26009.1 JAS31499.1 GEDC01029812 GEDC01012500 JAS07486.1 JAS24798.1 KZ288198 PBC33685.1 GL764129 EFZ18494.1 KQ978076 KYM97244.1 GBBI01003455 JAC15257.1 GBGD01000221 JAC88668.1 KQ981727 KYN37025.1 ACPB03021630 ACPB03021631 JH432011 GFAA01002157 JAU01278.1 GEFH01003251 JAP65330.1 GFPF01013403 MAA24549.1 GGLE01003844 MBY07970.1 GACK01001106 JAA63928.1 GEFM01004348 JAP71448.1 DS235068 EEB11252.1 GACK01001105 JAA63929.1 GEGO01001799 JAR93605.1 LNIX01000003 OXA58223.1 AMQN01002354 KB308962 ELT95953.1 KB631294 KB632328 ERL84216.1 ERL92540.1 ABLF02024336 ABLF02024337 GFXV01007058 MBW18863.1 GFAC01000400 JAT98788.1 GG666548 EEN57011.1 GGMS01016953 MBY86156.1 GECU01027990 JAS79716.1 NEDP02002474 OWF50762.1 HACG01025149 CEK72014.1 LJIJ01000549 ODM96342.1 KB201304 ESO97799.1 AHAT01005408 KK107088 EZA59763.1 GFDF01006402 JAV07682.1 GFDF01006403 JAV07681.1 GBEX01003378 JAI11182.1 PZQS01000003 PVD33410.1 JARO02001073 KPP76562.1 HADX01001351 SBP23583.1 CU896540 HADW01001341 SBP02741.1 JXUM01065830 KQ562367 KXJ76052.1 AXCN02000557 AXCP01008284 AXCP01008285 AEMK02000012 DQIR01021783 HCZ77258.1 JXLN01013336 KPM09271.1 HAEB01011202 SBQ57729.1 GFDL01002658 JAV32387.1 HAEG01001582 SBR65563.1
GDQN01010089 JAT80965.1 AGBW02014723 OWR41127.1 NWSH01005395 PCG64249.1 NWSH01007754 PCG62820.1 ODYU01010888 SOQ56274.1 ADTU01011884 PCG64250.1 NEVH01020353 PNF21369.1 GL452292 EFN77590.1 GBYB01010011 GBYB01010013 JAG79778.1 JAG79780.1 KQ435922 KOX68432.1 KQ414582 KOC70864.1 PNF21368.1 KQ434998 KZC13338.1 KQ976396 KYM92930.1 GEZM01087290 JAV58793.1 NNAY01003342 OXU19592.1 GL438684 EFN68621.1 QOIP01000008 RLU19823.1 KQ982934 KYQ49185.1 GEDC01011289 GEDC01005799 JAS26009.1 JAS31499.1 GEDC01029812 GEDC01012500 JAS07486.1 JAS24798.1 KZ288198 PBC33685.1 GL764129 EFZ18494.1 KQ978076 KYM97244.1 GBBI01003455 JAC15257.1 GBGD01000221 JAC88668.1 KQ981727 KYN37025.1 ACPB03021630 ACPB03021631 JH432011 GFAA01002157 JAU01278.1 GEFH01003251 JAP65330.1 GFPF01013403 MAA24549.1 GGLE01003844 MBY07970.1 GACK01001106 JAA63928.1 GEFM01004348 JAP71448.1 DS235068 EEB11252.1 GACK01001105 JAA63929.1 GEGO01001799 JAR93605.1 LNIX01000003 OXA58223.1 AMQN01002354 KB308962 ELT95953.1 KB631294 KB632328 ERL84216.1 ERL92540.1 ABLF02024336 ABLF02024337 GFXV01007058 MBW18863.1 GFAC01000400 JAT98788.1 GG666548 EEN57011.1 GGMS01016953 MBY86156.1 GECU01027990 JAS79716.1 NEDP02002474 OWF50762.1 HACG01025149 CEK72014.1 LJIJ01000549 ODM96342.1 KB201304 ESO97799.1 AHAT01005408 KK107088 EZA59763.1 GFDF01006402 JAV07682.1 GFDF01006403 JAV07681.1 GBEX01003378 JAI11182.1 PZQS01000003 PVD33410.1 JARO02001073 KPP76562.1 HADX01001351 SBP23583.1 CU896540 HADW01001341 SBP02741.1 JXUM01065830 KQ562367 KXJ76052.1 AXCN02000557 AXCP01008284 AXCP01008285 AEMK02000012 DQIR01021783 HCZ77258.1 JXLN01013336 KPM09271.1 HAEB01011202 SBQ57729.1 GFDL01002658 JAV32387.1 HAEG01001582 SBR65563.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000005205
+ More
UP000005203 UP000235965 UP000008237 UP000053105 UP000053825 UP000076502 UP000002358 UP000078540 UP000215335 UP000000311 UP000279307 UP000075809 UP000242457 UP000078542 UP000078541 UP000015103 UP000009046 UP000198287 UP000085678 UP000014760 UP000030742 UP000007819 UP000001554 UP000076420 UP000242188 UP000094527 UP000030746 UP000261540 UP000018468 UP000053097 UP000264820 UP000192224 UP000245119 UP000034805 UP000261440 UP000000437 UP000069940 UP000249989 UP000075886 UP000221080 UP000075880 UP000261400 UP000007635 UP000008227 UP000265120
UP000005203 UP000235965 UP000008237 UP000053105 UP000053825 UP000076502 UP000002358 UP000078540 UP000215335 UP000000311 UP000279307 UP000075809 UP000242457 UP000078542 UP000078541 UP000015103 UP000009046 UP000198287 UP000085678 UP000014760 UP000030742 UP000007819 UP000001554 UP000076420 UP000242188 UP000094527 UP000030746 UP000261540 UP000018468 UP000053097 UP000264820 UP000192224 UP000245119 UP000034805 UP000261440 UP000000437 UP000069940 UP000249989 UP000075886 UP000221080 UP000075880 UP000261400 UP000007635 UP000008227 UP000265120
Pfam
Interpro
IPR000195
Rab-GTPase-TBC_dom
+ More
IPR011993 PH-like_dom_sf
IPR036014 TCB1D9/TCB1D9B_PH-GRAM1
IPR036017 TCB1D9/TCB1D9B_PH-GRAM2
IPR035969 Rab-GTPase_TBC_sf
IPR004182 GRAM
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR018247 EF_Hand_1_Ca_BS
IPR005654 ATPase_AFG1-like
IPR027417 P-loop_NTPase
IPR034922 REX1-like_exo
IPR012337 RNaseH-like_sf
IPR013520 Exonuclease_RNaseT/DNA_pol3
IPR036397 RNaseH_sf
IPR001751 S100/CaBP-9k_CS
IPR001683 Phox
IPR001452 SH3_domain
IPR027267 AH/BAR_dom_sf
IPR036871 PX_dom_sf
IPR019497 Sorting_nexin_WASP-bd-dom
IPR036028 SH3-like_dom_sf
IPR011993 PH-like_dom_sf
IPR036014 TCB1D9/TCB1D9B_PH-GRAM1
IPR036017 TCB1D9/TCB1D9B_PH-GRAM2
IPR035969 Rab-GTPase_TBC_sf
IPR004182 GRAM
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR018247 EF_Hand_1_Ca_BS
IPR005654 ATPase_AFG1-like
IPR027417 P-loop_NTPase
IPR034922 REX1-like_exo
IPR012337 RNaseH-like_sf
IPR013520 Exonuclease_RNaseT/DNA_pol3
IPR036397 RNaseH_sf
IPR001751 S100/CaBP-9k_CS
IPR001683 Phox
IPR001452 SH3_domain
IPR027267 AH/BAR_dom_sf
IPR036871 PX_dom_sf
IPR019497 Sorting_nexin_WASP-bd-dom
IPR036028 SH3-like_dom_sf
SUPFAM
ProteinModelPortal
H9JBQ7
A0A194Q888
A0A194QQU1
A0A1E1W1V0
A0A212EI24
A0A2A4IX64
+ More
A0A2A4IUJ7 A0A2H1WV40 A0A158NCG7 A0A088AVK6 A0A2A4IYK2 A0A2J7PYF0 E2C358 A0A0C9RRM3 A0A0N0U388 A0A0L7RJ63 A0A2J7PYF6 A0A154PQ11 K7IPM7 A0A195BXB1 A0A1Y1KBA7 A0A232EMQ4 E2AD73 A0A3L8DI37 A0A151WNC2 A0A1B6E0Q9 A0A1B6C1U8 A0A2A3EPJ6 E9IM53 A0A151IBY4 A0A023F161 A0A069DX64 A0A195F8U1 T1HS16 T1JBL6 A0A1E1XPN5 A0A131XGW9 A0A224Z875 A0A2R5LEK8 L7MKW6 A0A131XYW0 E0VCZ6 L7ML78 A0A147BT99 A0A226EKI2 A0A1S3JSL2 R7TPZ1 A0A1S3JSN9 U4U0V3 J9K185 A0A2H8TX45 A0A1E1XHV0 C3YSA6 A0A2C9K5E8 A0A2S2R7Z9 A0A1B6HYF8 A0A210QPU5 A0A0B6ZTR9 A0A1D2MTT0 V4C7V3 A0A3B3T1T3 A0A3B3T105 A0A3B3T1Y9 W5N0E3 A0A026WUX3 A0A1L8DMK7 A0A1L8DMP1 A0A0F7Z5H4 A0A3B3T377 A0A3Q2XHJ3 A0A3B3T1E2 A0A3B3T320 A0A1W4YMM1 A0A2T7PJ41 A0A0P7V1N1 A0A1A7XZX8 A0A3B4EMN8 E7F7E8 A0A1W4YWN3 A0A1A7WB39 A0A286Y906 A0A182GID6 A0A182QVV2 A0A2D0SEL7 A0A182J9L3 A0A3B4ZMZ6 A0A2D0SDW3 A0A3B4DBT0 A0A3B3T167 G3PYY0 F1S448 A0A287B5J0 A0A132AE57 A0A3P8X003 A0A1A8FHX3 A0A1Q3FXT6 A0A1W3JAA4 A0A1A8N900
A0A2A4IUJ7 A0A2H1WV40 A0A158NCG7 A0A088AVK6 A0A2A4IYK2 A0A2J7PYF0 E2C358 A0A0C9RRM3 A0A0N0U388 A0A0L7RJ63 A0A2J7PYF6 A0A154PQ11 K7IPM7 A0A195BXB1 A0A1Y1KBA7 A0A232EMQ4 E2AD73 A0A3L8DI37 A0A151WNC2 A0A1B6E0Q9 A0A1B6C1U8 A0A2A3EPJ6 E9IM53 A0A151IBY4 A0A023F161 A0A069DX64 A0A195F8U1 T1HS16 T1JBL6 A0A1E1XPN5 A0A131XGW9 A0A224Z875 A0A2R5LEK8 L7MKW6 A0A131XYW0 E0VCZ6 L7ML78 A0A147BT99 A0A226EKI2 A0A1S3JSL2 R7TPZ1 A0A1S3JSN9 U4U0V3 J9K185 A0A2H8TX45 A0A1E1XHV0 C3YSA6 A0A2C9K5E8 A0A2S2R7Z9 A0A1B6HYF8 A0A210QPU5 A0A0B6ZTR9 A0A1D2MTT0 V4C7V3 A0A3B3T1T3 A0A3B3T105 A0A3B3T1Y9 W5N0E3 A0A026WUX3 A0A1L8DMK7 A0A1L8DMP1 A0A0F7Z5H4 A0A3B3T377 A0A3Q2XHJ3 A0A3B3T1E2 A0A3B3T320 A0A1W4YMM1 A0A2T7PJ41 A0A0P7V1N1 A0A1A7XZX8 A0A3B4EMN8 E7F7E8 A0A1W4YWN3 A0A1A7WB39 A0A286Y906 A0A182GID6 A0A182QVV2 A0A2D0SEL7 A0A182J9L3 A0A3B4ZMZ6 A0A2D0SDW3 A0A3B4DBT0 A0A3B3T167 G3PYY0 F1S448 A0A287B5J0 A0A132AE57 A0A3P8X003 A0A1A8FHX3 A0A1Q3FXT6 A0A1W3JAA4 A0A1A8N900
PDB
4P17
E-value=1.88007e-27,
Score=309
Ontologies
GO
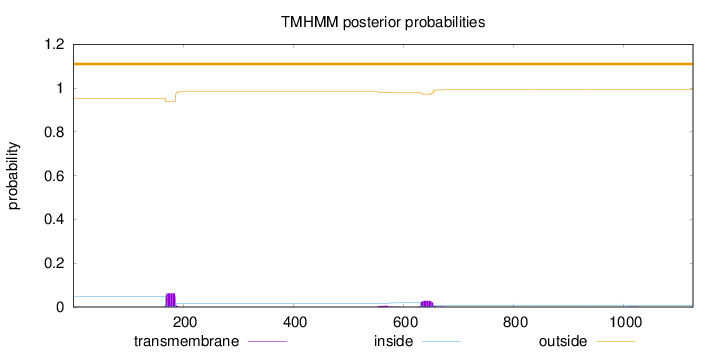
Topology
Length:
1126
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.88248
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.04753
outside
1 - 1126
Population Genetic Test Statistics
Pi
179.766318
Theta
175.227682
Tajima's D
0.657141
CLR
0
CSRT
0.552322383880806
Interpretation
Uncertain
