Pre Gene Modal
BGIBMGA006951
Annotation
hypothetical_protein_KGM_17238_[Danaus_plexippus]
Full name
Protein rogdi
Location in the cell
Cytoplasmic Reliability : 1.962 Nuclear Reliability : 1.37
Sequence
CDS
ATGGGTGACAACGAAAAAGAAGAAGCAGCTAATTTAAAAGAAGAGTTTGAATGGGTTCTTCGTGAAGAGGTTCATGCTATATTACACCAATTACACACAGTTTTAGTTGAATGTGCTCACAGGTTTCCTGTACCATTGTATGGTAATGAAGGCCAAAAACAGGATAAATTTATTTTGACATCACAACCTGAGCAGTTAAAGTGTATTGTGACCCTCACAGGAGATAGCATAACTCATGCAGATATTAGTTTCAAGGTCCTCCGTCAAATGCACACAATATGCCGTACCTCAATCAATCAAGATGGACCATGGAAACTTCAGCAGATACAAGATGCTGCAAACCATCTCCAACAAGCTATAGGCTATATTGATAATGTGGACAAACATTATGTTTTTCGATCATCAGAAGAAGTATTACATATAATTCAATGTCTTATTGGCTCTTTACAAAGAGCTCGGACTGCCTTAGTATTACCCAAAAAAAAAGCAATTGATGAGCTCATGAAGAGTCGAAATATGAAAGCTCTTTCACCAAATTTGCCTGAAGATTTAGCGATATCATTTTACATACAATCTCATAAATTGATATTTGTGGCGTACCATATTAGTTTTGTACATGGTACCATGAGGATTGATTCATGTCAAGCCGACTGTGCAGTGCCTTGGCTAAGTGATGTACTATTTATGCTGACAGCTGCATTACACATGTGTCAGCAATTAAAAGATAAGTTATGTGTTTTCTCGCAATACAAAGACTTCACAGTGGGATCAAGATCTGCCTCGATGGTGTTTAACTGA
Protein
MGDNEKEEAANLKEEFEWVLREEVHAILHQLHTVLVECAHRFPVPLYGNEGQKQDKFILTSQPEQLKCIVTLTGDSITHADISFKVLRQMHTICRTSINQDGPWKLQQIQDAANHLQQAIGYIDNVDKHYVFRSSEEVLHIIQCLIGSLQRARTALVLPKKKAIDELMKSRNMKALSPNLPEDLAISFYIQSHKLIFVAYHISFVHGTMRIDSCQADCAVPWLSDVLFMLTAALHMCQQLKDKLCVFSQYKDFTVGSRSASMVFN
Summary
Similarity
Belongs to the rogdi family.
Keywords
Alternative splicing
Complete proteome
Nucleus
Reference proteome
Feature
chain Protein rogdi
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2H1WZK2
A0A1E1WJG2
A0A194QWB6
A0A3S2MA04
H9JBQ6
A0A194Q8A0
+ More
A0A212EI13 A0A0N0PFE8 A0A182LK91 Q7PPB1 A0A182SBB5 A0A182WBL7 A0A182M2K3 A0A182K3E2 A0A182I4N1 A0A182X4N3 A7UUH2 A0A2M3ZIP4 A0A2M4BR16 A0A182VI07 A0A2M4BQ80 A0A2M4AU34 A0A2M4AU86 A0A182RDK5 A0A182NKJ0 A0A182J162 A0A182QHQ0 A0A1J1I7C2 A0A1Q3F0T3 A0A1Q3F0E6 A0A1L8DAX0 A0A1L8DAY3 W5JU02 A0A1L8DAY1 A0A182FD19 U5EZG1 A0A2J7PW06 A0A2J7PVZ9 A0A2J7PW07 B0W6V9 A0A182U792 A0A182PK11 A0A336MQA1 A0A182Y2F6 A0A1B0CKA7 A0A0K8TM40 A0A1L8EI54 A0A1S4EWQ3 T1PIP1 A0A1I8MLX6 Q17NC8 A0A067QN59 A0A1A9WFK1 A0A182GKD3 A0A1A9VHJ6 D3TQA4 A0A0A1X7R7 A0A1B0BFQ6 A0A2P8XQG1 A0A0K8WFR7 A0A1I8Q7G2 A0A1Y1LKY2 D6WZP6 B3M5H8 A0A1I8Q7J4 A0A1B6J284 A0A1B6GYE5 Q9VVE2 B4N3A0 B4PJY4 B3NDD3 B4HKA9 B4QNE9 Q9VVE2-2 A0A3B0KG16 A0A1W4W4K7 W8C4K8 E2BDB6 K7ITL6 B4LDT3 A0A232EZA2 A0A1L8E791 A0A1L8E6V7 B4KYE9 B4IZD7 A0A0Q9WV67 A0A0M3QX05 E9ILD5 A0A026VZ51 A0A195DYH2 F4WZL3 A0A195B7V1 A0A195FFF2 A0A158NGR6 A0A151IFD6 E1ZV53 A0A151XAC4 A0A0L0BWY9 A0A2A3EKA0 A0A087ZVT6
A0A212EI13 A0A0N0PFE8 A0A182LK91 Q7PPB1 A0A182SBB5 A0A182WBL7 A0A182M2K3 A0A182K3E2 A0A182I4N1 A0A182X4N3 A7UUH2 A0A2M3ZIP4 A0A2M4BR16 A0A182VI07 A0A2M4BQ80 A0A2M4AU34 A0A2M4AU86 A0A182RDK5 A0A182NKJ0 A0A182J162 A0A182QHQ0 A0A1J1I7C2 A0A1Q3F0T3 A0A1Q3F0E6 A0A1L8DAX0 A0A1L8DAY3 W5JU02 A0A1L8DAY1 A0A182FD19 U5EZG1 A0A2J7PW06 A0A2J7PVZ9 A0A2J7PW07 B0W6V9 A0A182U792 A0A182PK11 A0A336MQA1 A0A182Y2F6 A0A1B0CKA7 A0A0K8TM40 A0A1L8EI54 A0A1S4EWQ3 T1PIP1 A0A1I8MLX6 Q17NC8 A0A067QN59 A0A1A9WFK1 A0A182GKD3 A0A1A9VHJ6 D3TQA4 A0A0A1X7R7 A0A1B0BFQ6 A0A2P8XQG1 A0A0K8WFR7 A0A1I8Q7G2 A0A1Y1LKY2 D6WZP6 B3M5H8 A0A1I8Q7J4 A0A1B6J284 A0A1B6GYE5 Q9VVE2 B4N3A0 B4PJY4 B3NDD3 B4HKA9 B4QNE9 Q9VVE2-2 A0A3B0KG16 A0A1W4W4K7 W8C4K8 E2BDB6 K7ITL6 B4LDT3 A0A232EZA2 A0A1L8E791 A0A1L8E6V7 B4KYE9 B4IZD7 A0A0Q9WV67 A0A0M3QX05 E9ILD5 A0A026VZ51 A0A195DYH2 F4WZL3 A0A195B7V1 A0A195FFF2 A0A158NGR6 A0A151IFD6 E1ZV53 A0A151XAC4 A0A0L0BWY9 A0A2A3EKA0 A0A087ZVT6
Pubmed
26354079
19121390
22118469
20966253
12364791
14747013
+ More
17210077 20920257 23761445 25244985 26369729 17510324 25315136 24845553 26483478 20353571 25830018 29403074 28004739 18362917 19820115 17994087 10731132 12537572 12537569 17550304 22936249 24495485 20798317 20075255 28648823 21282665 24508170 30249741 21719571 21347285 26108605
17210077 20920257 23761445 25244985 26369729 17510324 25315136 24845553 26483478 20353571 25830018 29403074 28004739 18362917 19820115 17994087 10731132 12537572 12537569 17550304 22936249 24495485 20798317 20075255 28648823 21282665 24508170 30249741 21719571 21347285 26108605
EMBL
ODYU01012206
SOQ58386.1
GDQN01004053
JAT87001.1
KQ461175
KPJ07861.1
+ More
RSAL01000003 RVE54836.1 BABH01024523 KQ459302 KPJ01753.1 AGBW02014723 OWR41126.1 LADI01014852 KPJ20792.1 AAAB01008960 EAA10769.4 AXCM01003844 APCN01002332 EDO63950.1 GGFM01007663 MBW28414.1 GGFJ01006321 MBW55462.1 GGFJ01005940 MBW55081.1 GGFK01010962 MBW44283.1 GGFK01010961 MBW44282.1 AXCN02000396 CVRI01000043 CRK96181.1 GFDL01013884 JAV21161.1 GFDL01014019 JAV21026.1 GFDF01010463 JAV03621.1 GFDF01010466 JAV03618.1 ADMH02000378 ETN66773.1 GFDF01010465 JAV03619.1 GANO01000859 JAB59012.1 NEVH01020940 PNF20513.1 PNF20512.1 PNF20514.1 DS231850 EDS37105.1 UFQS01002022 UFQT01002022 SSX12995.1 SSX32436.1 AJWK01015949 AJWK01015950 AJWK01015951 GDAI01002179 JAI15424.1 GFDG01000464 JAV18335.1 KA647803 AFP62432.1 CH477200 EAT48207.1 KK853218 KDR09743.1 JXUM01013314 JXUM01013315 KQ560374 KXJ82730.1 EZ423606 ADD19882.1 GBXI01007105 JAD07187.1 JXJN01013628 PYGN01001530 PSN34247.1 GDHF01002303 JAI50011.1 GEZM01054669 JAV73498.1 KQ971372 EFA09669.1 CH902618 EDV39588.1 GECU01021759 GECU01014407 GECU01003661 JAS85947.1 JAS93299.1 JAT04046.1 GECZ01002326 JAS67443.1 AE014296 AY060379 BT023927 CH964062 EDW78839.1 CM000159 EDW94753.1 CH954178 EDV51995.1 CH480815 EDW41850.1 CM000363 CM002912 EDX10819.1 KMZ00191.1 OUUW01000009 SPP85269.1 GAMC01005444 JAC01112.1 GL447585 EFN86344.1 CH940647 EDW68956.1 NNAY01001503 OXU23772.1 GFDG01004226 JAV14573.1 GFDG01004328 JAV14471.1 CH933809 EDW17728.1 CH916366 EDV96692.1 KRF84101.1 CP012525 ALC45093.1 GL764074 EFZ18615.1 KK107681 QOIP01000006 EZA48109.1 RLU21512.1 KQ980107 KYN17654.1 GL888480 EGI60288.1 KQ976574 KYM80274.1 KQ981625 KYN39086.1 ADTU01015173 KQ977793 KYM99663.1 GL434441 EFN74912.1 KQ982339 KYQ57317.1 JRES01001303 KNC23754.1 KZ288219 PBC32233.1
RSAL01000003 RVE54836.1 BABH01024523 KQ459302 KPJ01753.1 AGBW02014723 OWR41126.1 LADI01014852 KPJ20792.1 AAAB01008960 EAA10769.4 AXCM01003844 APCN01002332 EDO63950.1 GGFM01007663 MBW28414.1 GGFJ01006321 MBW55462.1 GGFJ01005940 MBW55081.1 GGFK01010962 MBW44283.1 GGFK01010961 MBW44282.1 AXCN02000396 CVRI01000043 CRK96181.1 GFDL01013884 JAV21161.1 GFDL01014019 JAV21026.1 GFDF01010463 JAV03621.1 GFDF01010466 JAV03618.1 ADMH02000378 ETN66773.1 GFDF01010465 JAV03619.1 GANO01000859 JAB59012.1 NEVH01020940 PNF20513.1 PNF20512.1 PNF20514.1 DS231850 EDS37105.1 UFQS01002022 UFQT01002022 SSX12995.1 SSX32436.1 AJWK01015949 AJWK01015950 AJWK01015951 GDAI01002179 JAI15424.1 GFDG01000464 JAV18335.1 KA647803 AFP62432.1 CH477200 EAT48207.1 KK853218 KDR09743.1 JXUM01013314 JXUM01013315 KQ560374 KXJ82730.1 EZ423606 ADD19882.1 GBXI01007105 JAD07187.1 JXJN01013628 PYGN01001530 PSN34247.1 GDHF01002303 JAI50011.1 GEZM01054669 JAV73498.1 KQ971372 EFA09669.1 CH902618 EDV39588.1 GECU01021759 GECU01014407 GECU01003661 JAS85947.1 JAS93299.1 JAT04046.1 GECZ01002326 JAS67443.1 AE014296 AY060379 BT023927 CH964062 EDW78839.1 CM000159 EDW94753.1 CH954178 EDV51995.1 CH480815 EDW41850.1 CM000363 CM002912 EDX10819.1 KMZ00191.1 OUUW01000009 SPP85269.1 GAMC01005444 JAC01112.1 GL447585 EFN86344.1 CH940647 EDW68956.1 NNAY01001503 OXU23772.1 GFDG01004226 JAV14573.1 GFDG01004328 JAV14471.1 CH933809 EDW17728.1 CH916366 EDV96692.1 KRF84101.1 CP012525 ALC45093.1 GL764074 EFZ18615.1 KK107681 QOIP01000006 EZA48109.1 RLU21512.1 KQ980107 KYN17654.1 GL888480 EGI60288.1 KQ976574 KYM80274.1 KQ981625 KYN39086.1 ADTU01015173 KQ977793 KYM99663.1 GL434441 EFN74912.1 KQ982339 KYQ57317.1 JRES01001303 KNC23754.1 KZ288219 PBC32233.1
Proteomes
UP000053240
UP000283053
UP000005204
UP000053268
UP000007151
UP000075882
+ More
UP000007062 UP000075901 UP000075920 UP000075883 UP000075881 UP000075840 UP000076407 UP000075903 UP000075900 UP000075884 UP000075880 UP000075886 UP000183832 UP000000673 UP000069272 UP000235965 UP000002320 UP000075902 UP000075885 UP000076408 UP000092461 UP000095301 UP000008820 UP000027135 UP000091820 UP000069940 UP000249989 UP000078200 UP000092460 UP000245037 UP000095300 UP000007266 UP000007801 UP000000803 UP000007798 UP000002282 UP000008711 UP000001292 UP000000304 UP000268350 UP000192221 UP000008237 UP000002358 UP000008792 UP000215335 UP000009192 UP000001070 UP000092553 UP000053097 UP000279307 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000078542 UP000000311 UP000075809 UP000037069 UP000242457 UP000005203
UP000007062 UP000075901 UP000075920 UP000075883 UP000075881 UP000075840 UP000076407 UP000075903 UP000075900 UP000075884 UP000075880 UP000075886 UP000183832 UP000000673 UP000069272 UP000235965 UP000002320 UP000075902 UP000075885 UP000076408 UP000092461 UP000095301 UP000008820 UP000027135 UP000091820 UP000069940 UP000249989 UP000078200 UP000092460 UP000245037 UP000095300 UP000007266 UP000007801 UP000000803 UP000007798 UP000002282 UP000008711 UP000001292 UP000000304 UP000268350 UP000192221 UP000008237 UP000002358 UP000008792 UP000215335 UP000009192 UP000001070 UP000092553 UP000053097 UP000279307 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000078542 UP000000311 UP000075809 UP000037069 UP000242457 UP000005203
PRIDE
Pfam
Interpro
IPR028241
RAVE2/Rogdi
+ More
IPR001683 Phox
IPR036871 PX_dom_sf
IPR003114 Phox_assoc
IPR013937 Sorting_nexin_C
IPR018122 TF_fork_head_CS_1
IPR036390 WH_DNA-bd_sf
IPR000253 FHA_dom
IPR036388 WH-like_DNA-bd_sf
IPR008984 SMAD_FHA_dom_sf
IPR001766 Fork_head_dom
IPR030456 TF_fork_head_CS_2
IPR018247 EF_Hand_1_Ca_BS
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR001683 Phox
IPR036871 PX_dom_sf
IPR003114 Phox_assoc
IPR013937 Sorting_nexin_C
IPR018122 TF_fork_head_CS_1
IPR036390 WH_DNA-bd_sf
IPR000253 FHA_dom
IPR036388 WH-like_DNA-bd_sf
IPR008984 SMAD_FHA_dom_sf
IPR001766 Fork_head_dom
IPR030456 TF_fork_head_CS_2
IPR018247 EF_Hand_1_Ca_BS
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
Gene 3D
ProteinModelPortal
A0A2H1WZK2
A0A1E1WJG2
A0A194QWB6
A0A3S2MA04
H9JBQ6
A0A194Q8A0
+ More
A0A212EI13 A0A0N0PFE8 A0A182LK91 Q7PPB1 A0A182SBB5 A0A182WBL7 A0A182M2K3 A0A182K3E2 A0A182I4N1 A0A182X4N3 A7UUH2 A0A2M3ZIP4 A0A2M4BR16 A0A182VI07 A0A2M4BQ80 A0A2M4AU34 A0A2M4AU86 A0A182RDK5 A0A182NKJ0 A0A182J162 A0A182QHQ0 A0A1J1I7C2 A0A1Q3F0T3 A0A1Q3F0E6 A0A1L8DAX0 A0A1L8DAY3 W5JU02 A0A1L8DAY1 A0A182FD19 U5EZG1 A0A2J7PW06 A0A2J7PVZ9 A0A2J7PW07 B0W6V9 A0A182U792 A0A182PK11 A0A336MQA1 A0A182Y2F6 A0A1B0CKA7 A0A0K8TM40 A0A1L8EI54 A0A1S4EWQ3 T1PIP1 A0A1I8MLX6 Q17NC8 A0A067QN59 A0A1A9WFK1 A0A182GKD3 A0A1A9VHJ6 D3TQA4 A0A0A1X7R7 A0A1B0BFQ6 A0A2P8XQG1 A0A0K8WFR7 A0A1I8Q7G2 A0A1Y1LKY2 D6WZP6 B3M5H8 A0A1I8Q7J4 A0A1B6J284 A0A1B6GYE5 Q9VVE2 B4N3A0 B4PJY4 B3NDD3 B4HKA9 B4QNE9 Q9VVE2-2 A0A3B0KG16 A0A1W4W4K7 W8C4K8 E2BDB6 K7ITL6 B4LDT3 A0A232EZA2 A0A1L8E791 A0A1L8E6V7 B4KYE9 B4IZD7 A0A0Q9WV67 A0A0M3QX05 E9ILD5 A0A026VZ51 A0A195DYH2 F4WZL3 A0A195B7V1 A0A195FFF2 A0A158NGR6 A0A151IFD6 E1ZV53 A0A151XAC4 A0A0L0BWY9 A0A2A3EKA0 A0A087ZVT6
A0A212EI13 A0A0N0PFE8 A0A182LK91 Q7PPB1 A0A182SBB5 A0A182WBL7 A0A182M2K3 A0A182K3E2 A0A182I4N1 A0A182X4N3 A7UUH2 A0A2M3ZIP4 A0A2M4BR16 A0A182VI07 A0A2M4BQ80 A0A2M4AU34 A0A2M4AU86 A0A182RDK5 A0A182NKJ0 A0A182J162 A0A182QHQ0 A0A1J1I7C2 A0A1Q3F0T3 A0A1Q3F0E6 A0A1L8DAX0 A0A1L8DAY3 W5JU02 A0A1L8DAY1 A0A182FD19 U5EZG1 A0A2J7PW06 A0A2J7PVZ9 A0A2J7PW07 B0W6V9 A0A182U792 A0A182PK11 A0A336MQA1 A0A182Y2F6 A0A1B0CKA7 A0A0K8TM40 A0A1L8EI54 A0A1S4EWQ3 T1PIP1 A0A1I8MLX6 Q17NC8 A0A067QN59 A0A1A9WFK1 A0A182GKD3 A0A1A9VHJ6 D3TQA4 A0A0A1X7R7 A0A1B0BFQ6 A0A2P8XQG1 A0A0K8WFR7 A0A1I8Q7G2 A0A1Y1LKY2 D6WZP6 B3M5H8 A0A1I8Q7J4 A0A1B6J284 A0A1B6GYE5 Q9VVE2 B4N3A0 B4PJY4 B3NDD3 B4HKA9 B4QNE9 Q9VVE2-2 A0A3B0KG16 A0A1W4W4K7 W8C4K8 E2BDB6 K7ITL6 B4LDT3 A0A232EZA2 A0A1L8E791 A0A1L8E6V7 B4KYE9 B4IZD7 A0A0Q9WV67 A0A0M3QX05 E9ILD5 A0A026VZ51 A0A195DYH2 F4WZL3 A0A195B7V1 A0A195FFF2 A0A158NGR6 A0A151IFD6 E1ZV53 A0A151XAC4 A0A0L0BWY9 A0A2A3EKA0 A0A087ZVT6
PDB
5XQI
E-value=4.34975e-41,
Score=420
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Nucleus envelope
Nucleus envelope
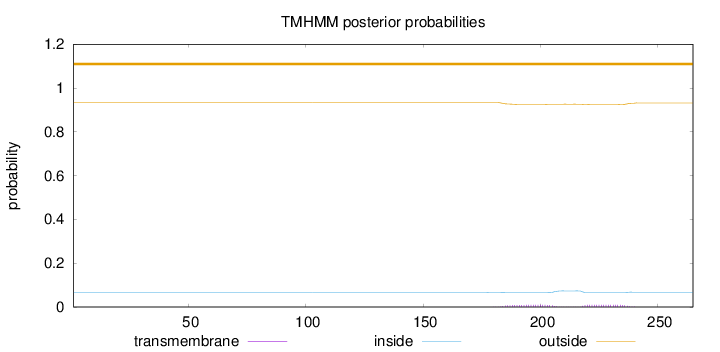
Length:
265
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.39201
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.06593
outside
1 - 265
Population Genetic Test Statistics
Pi
202.827719
Theta
195.043596
Tajima's D
0.499121
CLR
0
CSRT
0.514524273786311
Interpretation
Uncertain
