Gene
KWMTBOMO05830
Pre Gene Modal
BGIBMGA006906
Annotation
PREDICTED:_sorting_nexin-13-like_isoform_X2_[Bombyx_mori]
Full name
Sorting nexin-13
Location in the cell
Nuclear Reliability : 2.793
Sequence
CDS
ATGATAGCAAAGATTGCAGGATGGTTTGGTTTGATTAGCGTAATAATCACCGGTTTATTTGGAATTGGAATCTATTTTTCTTTACTGTTGTTTATTGTTTCTTTCATACTCGGTGTATTGACATTCCTGTATTGCGTTTATGATAACAAAAAAATATTTTCTGTTGATATAGACAGTCCTGATGATCCATTACAATGTAATAATTTCTCTATTACTCTTCCTAAGGTTCTAGAAGAGTTTCAAAAAAAGGTACCTTTATTAAAATTTGATAGTAGAATAACAGGAAGTGAAACTGTAGATTCATTTCTAAATGAAATAATTGTAATAATTATAAATGATTATGTTACACCATGGTATGAGATTTTGACAAATGATGATGAGTTCACTAAGCATGCAATAAAACGTTTGGTGCTTGCTACTGGTGCTAATGTAGTTAATAGGATAAAATGTGTGGATTGGATTAATTTGCTCTCAACAAGATTTCCTGAAGAACTAACAACTCATTTGAAGATGTTTAAGCAATCAAGAGTTCGCCTGAAACATGTAGAGTTAATGAATGCCAAAGAATCAGCAAATGATACTGCTAAATTAAGTCCAAAAAGGGATGAGAAGAAAACACACAGAAGGAATAAGAGTGAAACAGATTTATCTTGGTTACCAGACTCTCAAATATTTGGTAAATCAAAATTTTATGATAGCTCTGAAAATATTACAGGGAAAACATTAAAAGATATATTTTTTGAACTTGAATGTGCTATAGAAAATAAACAAATGTGTAGAGATGTGTTCTGTGATGATACAGATAAAGAATATGAGCTCCTTTGTGAAGTATCTGAAGCTATATTGTACTTGGTAGCTCCAGAAGAAGCATGGGGATGTCATGCAATGAAGACGATTTTGATTGATCTTTTATCAGCCGTTGTTTTACGCCCCATAATCAGTTTAATGAGTGACCCTGATAATATCAATAGAATGATCATAAAATCTTGTTGCAAAGATTCAATACTTCCATCAGAACTGTTTTTGATAGTCATAAGAAGCTGCGGGGATGCTGAAGAGTTGGACGCAACTTTAGAATTAGTAGAAAAGGATATACAGAAACTACATTCTAAAGACAGCGCCGGCGAATGGGAGCTTCAGGATCGTCAGAAATTATCCTCGTTACAGTACGTGACCAGGATAATACAGGCCAGAAGGGCTACGTTAGGTCCTCAAGAAGGTACTCTGAATAATTTGAATACACAAAAAATGTCCGAGGAAATTGCGAGAACATTGAAGTTTTTGAGAGCAACAGCAGCGTGGAAGTCTAACGCGCAATATTTACTAGAGATAGAGCTCCATGAATCAGACGCTCCGCACACATCGAAAGAGGTCATAGACAACTTACGTTCGTCCGCACTCGAGTTGTGCGATCTGTTTTTATCGCCGAACAAACCGCCGGTAATGGGCGTGCCAGAGAACTGTTATTCGGAACTCGTCCGTAAAATCACGACCGAAGGGGAGGAGTTTACGAACAATCCAGTTGTGTGTTTTGACGAGCTGCAGAAATGCGTCTGCGACGCTTTAGAGGACGATCCTTCTTGGCAAGCCGATTTTATGATGGAAACTGAGGATGGTACTTACAGAAGTAATAGTGACCTGAAAACCGATCCAAAATCGGACAGTAAAAAGTATAGCGGACCCTCAAGTTTGACAGCTAACTCCAGCTTGTCGATGTCGCACACGGAAATCAGTAGGAAAGCCAAAAATTTACTGTCTCCGCTATCGGCTTGTATCATAGAAACAGCACTAGTACAAGACAAAGGTCGAGCCTTTGGTATATACGCCATAGCCGTTACGAGACTTTCAGACAACGAGGTGTGGCACATATATAGGAGATACAGTGATTTCTATGACCTGCATTGCTCTATTAAAGAAAAGTGGCCAGAACTTGGGAACGTTCCATTTCCGGCTAAAAAGACGTTTCAGAATACGTCACGAGTAGTTTTGGAGAACAGGAAACGGATACTGAACGGCTACCTTCAGACTCTGGCCAGCCTTTCGAGGGAGGCAAAGTACATGGCGCTGCTGTCGCGGGACTACTTGGGGGGATTCCTCAGTCCGGTCAATCAAACCGAAAAACATAGTAATACGATCGATGCGCTTCTCGTGAACCCCTTGAGAGCCGGAATGAGGACATTGAAAAATATGCCGGATCAGTTCGTGAACACTGTCGACGGAGTTATGGACGGAATCTCAAAGGTCTTCCAAGGAAAGAACACTGAAAATTACTACGACGGCATTAAAACCGGCAACGCAACCGACGGACAAGACGACGGCGATGAAAGCATGGCATTGAAGCTCTTGGAAGAAGTGCTTGGCATCCGAGGGTTGTGGCTCAGGCGTCGACTTCTTGCGCCACTCAGAACCCTCATTGCGGATCGCGTGAACAGGAAAGTTTTAGAACTCGTATCGTCGTTGACGTCGCCTCGGAACGTGGTTCAGTATTTGAAAACGTTTAAGCAATGGTTAACTAGCCGACACAGCGCCAGTTACAGCGAAAGGGATGCGGCGACTAAAGCGAGAACTAGGGTTGCCGCTAAAGTTGCATTGCTCTCGGCCTGCTCTGATGATCTCAAACATATAGTCGGTACTGATTCAGCTCGAAGAGGTCTCCTCACAGTATTCGATTTATTCCAAATACAAGAAATCAACAGACGCTTAATATTCGTTTTGTTGGAAGTAACCCTAACCAATCTCTTTCCCGACAACAACATACAAGGTCTGTTCAAGAAACTTTACACGAATTCTCCTAGAGTATCAGCGCCGAAGAAGAATTCTGTTATATGA
Protein
MIAKIAGWFGLISVIITGLFGIGIYFSLLLFIVSFILGVLTFLYCVYDNKKIFSVDIDSPDDPLQCNNFSITLPKVLEEFQKKVPLLKFDSRITGSETVDSFLNEIIVIIINDYVTPWYEILTNDDEFTKHAIKRLVLATGANVVNRIKCVDWINLLSTRFPEELTTHLKMFKQSRVRLKHVELMNAKESANDTAKLSPKRDEKKTHRRNKSETDLSWLPDSQIFGKSKFYDSSENITGKTLKDIFFELECAIENKQMCRDVFCDDTDKEYELLCEVSEAILYLVAPEEAWGCHAMKTILIDLLSAVVLRPIISLMSDPDNINRMIIKSCCKDSILPSELFLIVIRSCGDAEELDATLELVEKDIQKLHSKDSAGEWELQDRQKLSSLQYVTRIIQARRATLGPQEGTLNNLNTQKMSEEIARTLKFLRATAAWKSNAQYLLEIELHESDAPHTSKEVIDNLRSSALELCDLFLSPNKPPVMGVPENCYSELVRKITTEGEEFTNNPVVCFDELQKCVCDALEDDPSWQADFMMETEDGTYRSNSDLKTDPKSDSKKYSGPSSLTANSSLSMSHTEISRKAKNLLSPLSACIIETALVQDKGRAFGIYAIAVTRLSDNEVWHIYRRYSDFYDLHCSIKEKWPELGNVPFPAKKTFQNTSRVVLENRKRILNGYLQTLASLSREAKYMALLSRDYLGGFLSPVNQTEKHSNTIDALLVNPLRAGMRTLKNMPDQFVNTVDGVMDGISKVFQGKNTENYYDGIKTGNATDGQDDGDESMALKLLEEVLGIRGLWLRRRLLAPLRTLIADRVNRKVLELVSSLTSPRNVVQYLKTFKQWLTSRHSASYSERDAATKARTRVAAKVALLSACSDDLKHIVGTDSARRGLLTVFDLFQIQEINRRLIFVLLEVTLTNLFPDNNIQGLFKKLYTNSPRVSAPKKNSVI
Summary
Description
May be involved in several stages of intracellular trafficking. Acts as a GAP for Galphas (By similarity). May play a role in endosome homeostasis.
Similarity
Belongs to the sorting nexin family.
Keywords
Complete proteome
Endosome
Lipid-binding
Membrane
Protein transport
Reference proteome
Signal transduction inhibitor
Transport
Feature
chain Sorting nexin-13
Uniprot
A0A1E1W2Y2
A0A194Q8M8
A0A212EI13
A0A2A4IU26
A0A194QSA1
A0A151IYW9
+ More
A0A0L7QSP7 A0A195CT69 A0A154PCQ2 A0A1B6DXD3 A0A0A9ZF17 A0A131YV03 A0A224Z721 L7MIL0 A0A131XPT3 V5I2C3 A0A131XFU5 A0A1E1XJW6 T1HE76 A0A1D2MYV4 B7PDH8 A0A2R5LGX5 C3YSL5 A0A2R2MTU3 A0A1B6BYU1 H0YWK9 U3K5M2 A0A218V7Y1 E1BZM0 E1C7R8 A0A1V4KC06 G1NCU0 F6WYZ9 M3YR94 A0A2U3VXM8 L5KLZ7 G9KQH1 A0A2Y9H7B6 A0A091VWD5 K7E145 G3T6H6 A0A2J8VEC7 A0A2U4ASA5 A0A2K5RG13 A0A341AUJ7 A0A2Y9PJ50 A0A2K5RG79 A0A341AUI2 A0A2K5RGA0 A0A093HXW7 G1LVT9 A0A2Y9DML8 A0A2U3XDJ5 A0A2U4AST1 A0A3Q7T273 A0A2Y9QXE8 U3INW5 W5PI30 K9J420 A0A1S3AGG7 A0A3Q2HPF8 A0A093EG06 A0A2Y9JCU4 A0A1S3AG65 A0A3Q7QKI3 A0A340XP54 A0A383YPY8 G1PPZ0 A0A340XVF7 M7AUF3 A0A3Q7WMP3 A0A3Q0G2N2 A0A3Q2XV67 A0A2D4ERR8 R0K4G3 A0A2D4ER02 A0A091VUX7 A0A3Q0D1I0 V9K7T7 A0A091Q4P8 A0A3Q1LUS6 A0A091HXX8 K1RAM8 Q6PHS6 E9QNG6 A0A2K6A199 A0A2K6A1C2 A0A0G2JUH8 V5H0X0 A0A2K6S375
A0A0L7QSP7 A0A195CT69 A0A154PCQ2 A0A1B6DXD3 A0A0A9ZF17 A0A131YV03 A0A224Z721 L7MIL0 A0A131XPT3 V5I2C3 A0A131XFU5 A0A1E1XJW6 T1HE76 A0A1D2MYV4 B7PDH8 A0A2R5LGX5 C3YSL5 A0A2R2MTU3 A0A1B6BYU1 H0YWK9 U3K5M2 A0A218V7Y1 E1BZM0 E1C7R8 A0A1V4KC06 G1NCU0 F6WYZ9 M3YR94 A0A2U3VXM8 L5KLZ7 G9KQH1 A0A2Y9H7B6 A0A091VWD5 K7E145 G3T6H6 A0A2J8VEC7 A0A2U4ASA5 A0A2K5RG13 A0A341AUJ7 A0A2Y9PJ50 A0A2K5RG79 A0A341AUI2 A0A2K5RGA0 A0A093HXW7 G1LVT9 A0A2Y9DML8 A0A2U3XDJ5 A0A2U4AST1 A0A3Q7T273 A0A2Y9QXE8 U3INW5 W5PI30 K9J420 A0A1S3AGG7 A0A3Q2HPF8 A0A093EG06 A0A2Y9JCU4 A0A1S3AG65 A0A3Q7QKI3 A0A340XP54 A0A383YPY8 G1PPZ0 A0A340XVF7 M7AUF3 A0A3Q7WMP3 A0A3Q0G2N2 A0A3Q2XV67 A0A2D4ERR8 R0K4G3 A0A2D4ER02 A0A091VUX7 A0A3Q0D1I0 V9K7T7 A0A091Q4P8 A0A3Q1LUS6 A0A091HXX8 K1RAM8 Q6PHS6 E9QNG6 A0A2K6A199 A0A2K6A1C2 A0A0G2JUH8 V5H0X0 A0A2K6S375
Pubmed
26354079
22118469
25401762
26823975
26830274
28797301
+ More
25576852 25765539 28049606 29209593 27289101 18563158 26383154 20360741 15592404 20838655 19892987 23258410 23236062 17495919 20010809 23749191 20809919 21993624 23624526 26319212 28071753 24402279 19393038 22992520 15489334 17077144 19468303 15057822
25576852 25765539 28049606 29209593 27289101 18563158 26383154 20360741 15592404 20838655 19892987 23258410 23236062 17495919 20010809 23749191 20809919 21993624 23624526 26319212 28071753 24402279 19393038 22992520 15489334 17077144 19468303 15057822
EMBL
GDQN01009724
JAT81330.1
KQ459302
KPJ01754.1
AGBW02014723
OWR41126.1
+ More
NWSH01007589 PCG62898.1 KQ461175 KPJ07860.1 KQ980735 KYN13761.1 KQ414756 KOC61655.1 KQ977305 KYN03717.1 KQ434868 KZC09194.1 GEDC01006960 JAS30338.1 GBHO01001606 GDHC01004069 JAG41998.1 JAQ14560.1 GEDV01006247 JAP82310.1 GFPF01011985 MAA23131.1 GACK01001941 JAA63093.1 GEFM01006573 JAP69223.1 GANP01000831 JAB83637.1 GEFH01002558 JAP66023.1 GFAA01003824 JAT99610.1 ACPB03024690 ACPB03024691 ACPB03024692 LJIJ01000375 ODM98180.1 ABJB010255224 ABJB010380668 ABJB010395578 ABJB010621187 ABJB011073047 DS689861 EEC04650.1 GGLE01004654 MBY08780.1 GG666549 EEN56716.1 GEDC01030880 JAS06418.1 ABQF01013497 ABQF01013498 ABQF01013499 AGTO01020330 MUZQ01000031 OWK62093.1 AADN05000033 LSYS01003958 OPJ81891.1 AEYP01012994 AEYP01012995 AEYP01012996 AEYP01012997 KB030660 ELK12560.1 JP018552 AES07150.1 KL410115 KFQ93953.1 NDHI03003421 PNJ55876.1 KL206634 KFV84355.1 ACTA01081149 ACTA01089149 ACTA01097149 ACTA01105149 ACTA01113149 ADON01042468 ADON01042469 AMGL01086205 AMGL01086206 AMGL01086207 AMGL01086208 GABZ01000509 JAA53016.1 KL243778 KFV13456.1 AAPE02008954 AAPE02008955 AAPE02008956 KB577554 EMP26615.1 IACJ01013648 LAA37636.1 KB742773 EOB04632.1 IACJ01013651 LAA37643.1 KK734334 KFR06318.1 JW861622 AFO94139.1 KK685868 KFQ14521.1 KL218015 KFP01154.1 JH816193 EKC38290.1 BC056394 AC122314 AC140376 AABR07063894 AABR07063895 GANP01013924 JAB70544.1
NWSH01007589 PCG62898.1 KQ461175 KPJ07860.1 KQ980735 KYN13761.1 KQ414756 KOC61655.1 KQ977305 KYN03717.1 KQ434868 KZC09194.1 GEDC01006960 JAS30338.1 GBHO01001606 GDHC01004069 JAG41998.1 JAQ14560.1 GEDV01006247 JAP82310.1 GFPF01011985 MAA23131.1 GACK01001941 JAA63093.1 GEFM01006573 JAP69223.1 GANP01000831 JAB83637.1 GEFH01002558 JAP66023.1 GFAA01003824 JAT99610.1 ACPB03024690 ACPB03024691 ACPB03024692 LJIJ01000375 ODM98180.1 ABJB010255224 ABJB010380668 ABJB010395578 ABJB010621187 ABJB011073047 DS689861 EEC04650.1 GGLE01004654 MBY08780.1 GG666549 EEN56716.1 GEDC01030880 JAS06418.1 ABQF01013497 ABQF01013498 ABQF01013499 AGTO01020330 MUZQ01000031 OWK62093.1 AADN05000033 LSYS01003958 OPJ81891.1 AEYP01012994 AEYP01012995 AEYP01012996 AEYP01012997 KB030660 ELK12560.1 JP018552 AES07150.1 KL410115 KFQ93953.1 NDHI03003421 PNJ55876.1 KL206634 KFV84355.1 ACTA01081149 ACTA01089149 ACTA01097149 ACTA01105149 ACTA01113149 ADON01042468 ADON01042469 AMGL01086205 AMGL01086206 AMGL01086207 AMGL01086208 GABZ01000509 JAA53016.1 KL243778 KFV13456.1 AAPE02008954 AAPE02008955 AAPE02008956 KB577554 EMP26615.1 IACJ01013648 LAA37636.1 KB742773 EOB04632.1 IACJ01013651 LAA37643.1 KK734334 KFR06318.1 JW861622 AFO94139.1 KK685868 KFQ14521.1 KL218015 KFP01154.1 JH816193 EKC38290.1 BC056394 AC122314 AC140376 AABR07063894 AABR07063895 GANP01013924 JAB70544.1
Proteomes
UP000053268
UP000007151
UP000218220
UP000053240
UP000078492
UP000053825
+ More
UP000078542 UP000076502 UP000015103 UP000094527 UP000001555 UP000001554 UP000085678 UP000007754 UP000016665 UP000197619 UP000000539 UP000190648 UP000001645 UP000002281 UP000000715 UP000245340 UP000010552 UP000248481 UP000053283 UP000002280 UP000007646 UP000245320 UP000233040 UP000252040 UP000248483 UP000053584 UP000008912 UP000248480 UP000245341 UP000286640 UP000016666 UP000002356 UP000079721 UP000248482 UP000286641 UP000265300 UP000001074 UP000031443 UP000286642 UP000189705 UP000264820 UP000053605 UP000189706 UP000009136 UP000054308 UP000005408 UP000000589 UP000233140 UP000002494 UP000233220
UP000078542 UP000076502 UP000015103 UP000094527 UP000001555 UP000001554 UP000085678 UP000007754 UP000016665 UP000197619 UP000000539 UP000190648 UP000001645 UP000002281 UP000000715 UP000245340 UP000010552 UP000248481 UP000053283 UP000002280 UP000007646 UP000245320 UP000233040 UP000252040 UP000248483 UP000053584 UP000008912 UP000248480 UP000245341 UP000286640 UP000016666 UP000002356 UP000079721 UP000248482 UP000286641 UP000265300 UP000001074 UP000031443 UP000286642 UP000189705 UP000264820 UP000053605 UP000189706 UP000009136 UP000054308 UP000005408 UP000000589 UP000233140 UP000002494 UP000233220
Interpro
Gene 3D
ProteinModelPortal
A0A1E1W2Y2
A0A194Q8M8
A0A212EI13
A0A2A4IU26
A0A194QSA1
A0A151IYW9
+ More
A0A0L7QSP7 A0A195CT69 A0A154PCQ2 A0A1B6DXD3 A0A0A9ZF17 A0A131YV03 A0A224Z721 L7MIL0 A0A131XPT3 V5I2C3 A0A131XFU5 A0A1E1XJW6 T1HE76 A0A1D2MYV4 B7PDH8 A0A2R5LGX5 C3YSL5 A0A2R2MTU3 A0A1B6BYU1 H0YWK9 U3K5M2 A0A218V7Y1 E1BZM0 E1C7R8 A0A1V4KC06 G1NCU0 F6WYZ9 M3YR94 A0A2U3VXM8 L5KLZ7 G9KQH1 A0A2Y9H7B6 A0A091VWD5 K7E145 G3T6H6 A0A2J8VEC7 A0A2U4ASA5 A0A2K5RG13 A0A341AUJ7 A0A2Y9PJ50 A0A2K5RG79 A0A341AUI2 A0A2K5RGA0 A0A093HXW7 G1LVT9 A0A2Y9DML8 A0A2U3XDJ5 A0A2U4AST1 A0A3Q7T273 A0A2Y9QXE8 U3INW5 W5PI30 K9J420 A0A1S3AGG7 A0A3Q2HPF8 A0A093EG06 A0A2Y9JCU4 A0A1S3AG65 A0A3Q7QKI3 A0A340XP54 A0A383YPY8 G1PPZ0 A0A340XVF7 M7AUF3 A0A3Q7WMP3 A0A3Q0G2N2 A0A3Q2XV67 A0A2D4ERR8 R0K4G3 A0A2D4ER02 A0A091VUX7 A0A3Q0D1I0 V9K7T7 A0A091Q4P8 A0A3Q1LUS6 A0A091HXX8 K1RAM8 Q6PHS6 E9QNG6 A0A2K6A199 A0A2K6A1C2 A0A0G2JUH8 V5H0X0 A0A2K6S375
A0A0L7QSP7 A0A195CT69 A0A154PCQ2 A0A1B6DXD3 A0A0A9ZF17 A0A131YV03 A0A224Z721 L7MIL0 A0A131XPT3 V5I2C3 A0A131XFU5 A0A1E1XJW6 T1HE76 A0A1D2MYV4 B7PDH8 A0A2R5LGX5 C3YSL5 A0A2R2MTU3 A0A1B6BYU1 H0YWK9 U3K5M2 A0A218V7Y1 E1BZM0 E1C7R8 A0A1V4KC06 G1NCU0 F6WYZ9 M3YR94 A0A2U3VXM8 L5KLZ7 G9KQH1 A0A2Y9H7B6 A0A091VWD5 K7E145 G3T6H6 A0A2J8VEC7 A0A2U4ASA5 A0A2K5RG13 A0A341AUJ7 A0A2Y9PJ50 A0A2K5RG79 A0A341AUI2 A0A2K5RGA0 A0A093HXW7 G1LVT9 A0A2Y9DML8 A0A2U3XDJ5 A0A2U4AST1 A0A3Q7T273 A0A2Y9QXE8 U3INW5 W5PI30 K9J420 A0A1S3AGG7 A0A3Q2HPF8 A0A093EG06 A0A2Y9JCU4 A0A1S3AG65 A0A3Q7QKI3 A0A340XP54 A0A383YPY8 G1PPZ0 A0A340XVF7 M7AUF3 A0A3Q7WMP3 A0A3Q0G2N2 A0A3Q2XV67 A0A2D4ERR8 R0K4G3 A0A2D4ER02 A0A091VUX7 A0A3Q0D1I0 V9K7T7 A0A091Q4P8 A0A3Q1LUS6 A0A091HXX8 K1RAM8 Q6PHS6 E9QNG6 A0A2K6A199 A0A2K6A1C2 A0A0G2JUH8 V5H0X0 A0A2K6S375
PDB
6EE0
E-value=1.44669e-06,
Score=128
Ontologies
GO
Topology
Subcellular location
Early endosome membrane
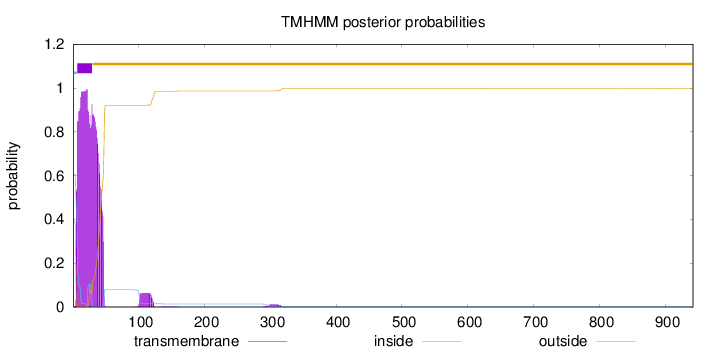
Length:
942
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
36.2224900000001
Exp number, first 60 AAs:
34.62796
Total prob of N-in:
0.60846
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 942
Population Genetic Test Statistics
Pi
272.632054
Theta
176.686328
Tajima's D
1.656712
CLR
0.645835
CSRT
0.818909054547273
Interpretation
Uncertain
