Gene
KWMTBOMO05818 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006908
Annotation
PREDICTED:_low-density_lipoprotein_receptor-related_protein_2_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 4.181
Sequence
CDS
ATGAACGTCAAATCAAGCACTATAATGTACTTACTATGGTTCGCCTTATTTATTGTGATGATTTCACAAGCCGATGCGTCAATATTGGGTTCTCATTGTCGAAGCAATGCGGACTGTAAAGTGGCCTATGCTCATTGTGACAGAGGAATGTGCATGTGTCAGCCCTATTACGCTCGCGTTGATAACTCCACCTGCTTGGAATCAACTCTACTTGGTTCAGAGTGCATGGTGGCTGAACAGTGCACGCAGAAAGTGGCTTACAGCGCATGCTTGGAAGGCGTTTGCCGTTGTACAGACGGACACCTACAATTTAGAAAACACACTTGCCTATCCCCTGCTCCGCCTGGTCACGTTTGCTACAGTAATGAACATTGTCGGTTATGGGATAAGGAAAGCCGATGCGAATTCGTGATTCTTAATCTATTCGGGAGATGCGCTTGCAATTCCTACTATAAACAAAATGGCGATAAATGTGTGCTCTCGAAGATTTCCCCCTATGCAACCGAAGCAGTGATCGCAGAACAAGATTCCGCTCTGAGTCCTATGAATGGCCCTCAAAATACGATGAATAGTAATGGCCAGACTTCCTTCAGCGCTGAGTCAGGATTTAATGAGAGAAACCAGGCTCGACCAAGCAAAACTCCCATATTTTCCGATAATGATGTTTTGAGTACAGAAGACGATAATCTATTAATGCAAGCCGTATTGAAGGTGCCAACTATAGAACCAGCCAAAAAATTACCTGTCTCTAACAGAAAAGAGGGCGAACCAAATGAGACAGTTGGTGTTAATGAAATGACTGCAAACTTGCATGCAAACGATCAGCCTGTTTACAGGGTTGTGACGCAACCAACAGTAGATAAAAAGTCTAATAAGAAGACAGGGTCCACAAAAGAAACTTCAGAAGGTAAACCGCACAAAACTGGACATCAAAAAAAATCATCTCTCAAGGATAAGCAACGCATTCGCACTGATGTCTCTGGCGACCTTGGACCTTCATCTCTAGGTATGCCTTGTATGACAGACGCACAATGCCATCTAGTGGATCCTCATACGAAATGCTTAAACAAACGATGTGATTGCGCTCATCGCACTAATTCGACATCAGCTTGTTCGGCTCGCAACAGGGGCTGCTTACCTGGAACATTTCAATGTCGGTCTACGGGCACCTGCATAAGCTGGTACTTCGTCTGTGATGGGCGTAAGGATTGTTCTGATGGATCCGATGAGAAGTGTCAAGATGGGACGGGCGAGCGGTGTCCTATGAACTCGTTCAGATGTGGGGGTCCTGGTTCGCCGTGTGTATCAAGGGCGGCCAGGTGTGATGGCACTCCACAGTGTCCCGGAGGCGAAGATGAAAGAAACTGTAAGGCGACGAGACGCAGGGGATGTCCCCGTCACACGTTCCGTTGCGGTTCTGGTGAATGTTTACCGGAGTACGAGTTTTGTAACGCTATAATATCATGCAAAGACGCTTCTGATGAACCACCACATTTATGTACAGAACAATCAAGATGGCGAGCCGCTGATTTCTGCCCTCTTCGATGTGGCAATGGCCGTTGCAGAAGCACAGCCGTAGCATGTTCAGGACGCGACGGGTGTGGAGATAACAGCGATGAAATTGCATGTTCTGTTTGTAAGTGTCCATCGCCTCAGACGCCTTGA
Protein
MNVKSSTIMYLLWFALFIVMISQADASILGSHCRSNADCKVAYAHCDRGMCMCQPYYARVDNSTCLESTLLGSECMVAEQCTQKVAYSACLEGVCRCTDGHLQFRKHTCLSPAPPGHVCYSNEHCRLWDKESRCEFVILNLFGRCACNSYYKQNGDKCVLSKISPYATEAVIAEQDSALSPMNGPQNTMNSNGQTSFSAESGFNERNQARPSKTPIFSDNDVLSTEDDNLLMQAVLKVPTIEPAKKLPVSNRKEGEPNETVGVNEMTANLHANDQPVYRVVTQPTVDKKSNKKTGSTKETSEGKPHKTGHQKKSSLKDKQRIRTDVSGDLGPSSLGMPCMTDAQCHLVDPHTKCLNKRCDCAHRTNSTSACSARNRGCLPGTFQCRSTGTCISWYFVCDGRKDCSDGSDEKCQDGTGERCPMNSFRCGGPGSPCVSRAARCDGTPQCPGGEDERNCKATRRRGCPRHTFRCGSGECLPEYEFCNAIISCKDASDEPPHLCTEQSRWRAADFCPLRCGNGRCRSTAVACSGRDGCGDNSDEIACSVCKCPSPQTP
Summary
Uniprot
H9JBL3
A0A2A4JS82
A0A194QS91
A0A194Q8B1
A0A212F110
A0A0L7LLJ7
+ More
A0A158NPH2 A0A195E658 A0A151WR70 A0A026WH94 A0A195EUY8 K7IZ30 A0A0C9RG06 A0A195BA34 E2C919 E2ACT5 F4WNZ7 A0A195CP09 A0A067RA40 A0A1B6DKZ2 A0A0N0BJK5 A0A310S979 A0A2S2PS21 A0A087ZWI8 A0A2S2QTF5 X1WJM5 A0A154P0J9 A0A0A1XA81 A0A182Q8N0 A0A1J1ITD7 A0A0L0CR37 A0A0L7QVK5
A0A158NPH2 A0A195E658 A0A151WR70 A0A026WH94 A0A195EUY8 K7IZ30 A0A0C9RG06 A0A195BA34 E2C919 E2ACT5 F4WNZ7 A0A195CP09 A0A067RA40 A0A1B6DKZ2 A0A0N0BJK5 A0A310S979 A0A2S2PS21 A0A087ZWI8 A0A2S2QTF5 X1WJM5 A0A154P0J9 A0A0A1XA81 A0A182Q8N0 A0A1J1ITD7 A0A0L0CR37 A0A0L7QVK5
Pubmed
EMBL
BABH01024481
BABH01024482
NWSH01000730
PCG74548.1
KQ461175
KPJ07850.1
+ More
KQ459302 KPJ01763.1 AGBW02011011 OWR47391.1 JTDY01000658 KOB76300.1 ADTU01022544 ADTU01022545 KQ979608 KYN20334.1 KQ982807 KYQ50399.1 KK107207 EZA55432.1 KQ981965 KYN31707.1 GBYB01007260 JAG77027.1 KQ976542 KYM81090.1 GL453768 EFN75555.1 GL438582 EFN68733.1 GL888243 EGI63928.1 KQ977574 KYN01844.1 KK852824 KDR15449.1 GEDC01010976 JAS26322.1 KQ435713 KOX79262.1 KQ763441 OAD55110.1 GGMR01019509 MBY32128.1 GGMS01011209 MBY80412.1 ABLF02017102 KQ434792 KZC05445.1 GBXI01006652 JAD07640.1 AXCN02000398 CVRI01000059 CRL03471.1 JRES01000036 KNC34711.1 KQ414727 KOC62589.1
KQ459302 KPJ01763.1 AGBW02011011 OWR47391.1 JTDY01000658 KOB76300.1 ADTU01022544 ADTU01022545 KQ979608 KYN20334.1 KQ982807 KYQ50399.1 KK107207 EZA55432.1 KQ981965 KYN31707.1 GBYB01007260 JAG77027.1 KQ976542 KYM81090.1 GL453768 EFN75555.1 GL438582 EFN68733.1 GL888243 EGI63928.1 KQ977574 KYN01844.1 KK852824 KDR15449.1 GEDC01010976 JAS26322.1 KQ435713 KOX79262.1 KQ763441 OAD55110.1 GGMR01019509 MBY32128.1 GGMS01011209 MBY80412.1 ABLF02017102 KQ434792 KZC05445.1 GBXI01006652 JAD07640.1 AXCN02000398 CVRI01000059 CRL03471.1 JRES01000036 KNC34711.1 KQ414727 KOC62589.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000005205 UP000078492 UP000075809 UP000053097 UP000078541 UP000002358 UP000078540 UP000008237 UP000000311 UP000007755 UP000078542 UP000027135 UP000053105 UP000005203 UP000007819 UP000076502 UP000075886 UP000183832 UP000037069 UP000053825
UP000005205 UP000078492 UP000075809 UP000053097 UP000078541 UP000002358 UP000078540 UP000008237 UP000000311 UP000007755 UP000078542 UP000027135 UP000053105 UP000005203 UP000007819 UP000076502 UP000075886 UP000183832 UP000037069 UP000053825
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JBL3
A0A2A4JS82
A0A194QS91
A0A194Q8B1
A0A212F110
A0A0L7LLJ7
+ More
A0A158NPH2 A0A195E658 A0A151WR70 A0A026WH94 A0A195EUY8 K7IZ30 A0A0C9RG06 A0A195BA34 E2C919 E2ACT5 F4WNZ7 A0A195CP09 A0A067RA40 A0A1B6DKZ2 A0A0N0BJK5 A0A310S979 A0A2S2PS21 A0A087ZWI8 A0A2S2QTF5 X1WJM5 A0A154P0J9 A0A0A1XA81 A0A182Q8N0 A0A1J1ITD7 A0A0L0CR37 A0A0L7QVK5
A0A158NPH2 A0A195E658 A0A151WR70 A0A026WH94 A0A195EUY8 K7IZ30 A0A0C9RG06 A0A195BA34 E2C919 E2ACT5 F4WNZ7 A0A195CP09 A0A067RA40 A0A1B6DKZ2 A0A0N0BJK5 A0A310S979 A0A2S2PS21 A0A087ZWI8 A0A2S2QTF5 X1WJM5 A0A154P0J9 A0A0A1XA81 A0A182Q8N0 A0A1J1ITD7 A0A0L0CR37 A0A0L7QVK5
PDB
3M0C
E-value=1.14669e-16,
Score=213
Ontologies
GO
PANTHER
Topology
SignalP
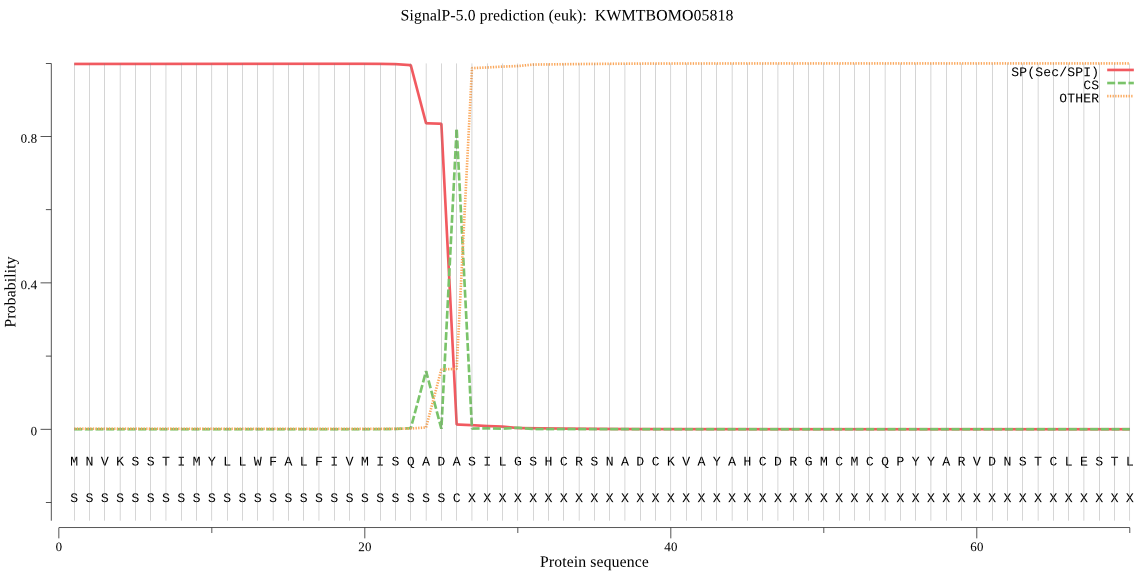
Position: 1 - 26,
Likelihood: 0.998778
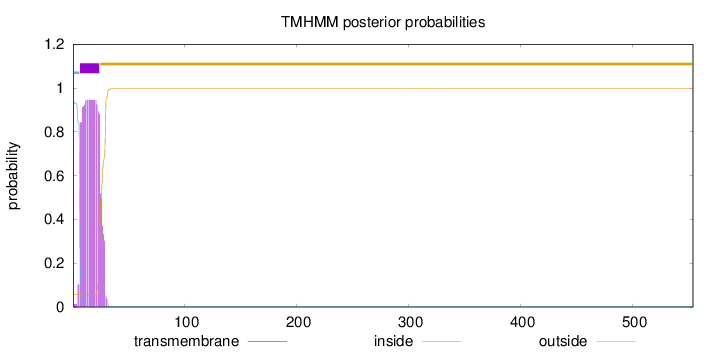
Length:
554
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.96415
Exp number, first 60 AAs:
18.96399
Total prob of N-in:
0.94332
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 24
outside
25 - 554
Population Genetic Test Statistics
Pi
162.135056
Theta
160.029072
Tajima's D
0.252782
CLR
0.712795
CSRT
0.444827758612069
Interpretation
Uncertain
