Pre Gene Modal
BGIBMGA006908
Annotation
PREDICTED:_uncharacterized_protein_LOC101735991_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.404
Sequence
CDS
ATGATTGGTTTGGATTCGATGTCTTTGCCAAGACTAGCGCAGACAATGACACGAACAATCGACCATCTATACGAAAATTTCTGGTTGCTTTTCAAAGGATATCATAAGGTTGAAGATAATACATTTCCAAATCTCATGGCGGTATTGACTGGAAAAAACTTGACGGCAATTACAAATTTATGTACGAATAGAATGGATCGATGCAATGAAATGATGATATGGAGCGACTTCAAACGTGACGGTTATATAACAGCTTACGGTGAAGACTACCTGCGTTTGCCGGATACTTTCACAAACAGATATGCTTATAATAATTCGCCCACCGACCACTATCTGAGGCCATTTTTTCTTCAGGGTCAGAATCCGCAAAGTAAGGGGCTTATTAACAAAGCTACTTTATGTGGGATGTTTATGGATCCTCTTTACTAG
Protein
MIGLDSMSLPRLAQTMTRTIDHLYENFWLLFKGYHKVEDNTFPNLMAVLTGKNLTAITNLCTNRMDRCNEMMIWSDFKRDGYITAYGEDYLRLPDTFTNRYAYNNSPTDHYLRPFFLQGQNPQSKGLINKATLCGMFMDPLY
Summary
Uniprot
H9JBL3
A0A2H1VLI2
A0A2A4JRV5
A0A212EUQ7
A0A194QS91
A0A194Q8N7
+ More
A0A212F0X9 A0A067R7H4 A0A0A9ZE12 A0A1B6J3R1 A0A2J7QJD5 A0A146LTH0 B4KYD4 A0A2J7QJF3 A0A2J7QJG0 T1HQF2 M9PEJ0 A0A0R1DYK0 A0A0R1E097 B4PG32 A0A1B0FM17 A0A084WRY1 A0A0Q5U387 A0A0A9YH28 B3NFS7 A0A182MPD0 A0A0J9UH40 A0A0J9RRE0 A0A182J4D0 A0A182P0X0 Q16GN7 A0A0P6IX53 A0A2R7X6X6 A0A182GTW9 A0A182Y378 A0A1B6C907 A0A1S4G1C9 B0VZL8 Q9VSJ3 Q7Q631 A0A182JS97 A0A182VP48 A7UU09 A0A182Q8B6 A0A336LIZ0 A0A139WMR3 A0A182RXH5 A0A2M4AJW8 B4KYD6 A0A182NKH8 A0A1W4US11 A0A1W4UF61 A0A1W4UEM4 A0A1B6LF10 W8BP58 A0A0K8U1F0 W8B9F7 A0A0A1X6X0 A0A1A9VYN0 B4K3H9 B4IZF0 B4IZE9 B4LDR7 A0A0Q9WTU0 B4LDR8 A0A2M4AGQ7 A0A0Q9WHL4 A0A2M3ZI29 A0A1B0CTD7 A0A0N0BJJ8 A0A034VIR4 A0A0K8URL1 A0A182FD32 A0A1I8MYF3 A0A034VH53 A0A0K8U8R6 A0A0K8VB69 W5JL99 A0A0L0CQQ8 A0A1J1ISV5 A0A1I8P764 A0A182WHQ3 A0A0N8P0U5 B3M5J1 A0A1A9WTF2 A0A3B0JT53 T1GHE2 A0A0M4ED41 B4KYD5 B4H633 A0A0Q9XP79 A0A1B6F5X8 A0A1B6EWE6 A0A3B0JT52 A0A3B0KN97 A0A0M4EB32 A0A3B0JT36 A0A3B0KHR6 B4JUE7
A0A212F0X9 A0A067R7H4 A0A0A9ZE12 A0A1B6J3R1 A0A2J7QJD5 A0A146LTH0 B4KYD4 A0A2J7QJF3 A0A2J7QJG0 T1HQF2 M9PEJ0 A0A0R1DYK0 A0A0R1E097 B4PG32 A0A1B0FM17 A0A084WRY1 A0A0Q5U387 A0A0A9YH28 B3NFS7 A0A182MPD0 A0A0J9UH40 A0A0J9RRE0 A0A182J4D0 A0A182P0X0 Q16GN7 A0A0P6IX53 A0A2R7X6X6 A0A182GTW9 A0A182Y378 A0A1B6C907 A0A1S4G1C9 B0VZL8 Q9VSJ3 Q7Q631 A0A182JS97 A0A182VP48 A7UU09 A0A182Q8B6 A0A336LIZ0 A0A139WMR3 A0A182RXH5 A0A2M4AJW8 B4KYD6 A0A182NKH8 A0A1W4US11 A0A1W4UF61 A0A1W4UEM4 A0A1B6LF10 W8BP58 A0A0K8U1F0 W8B9F7 A0A0A1X6X0 A0A1A9VYN0 B4K3H9 B4IZF0 B4IZE9 B4LDR7 A0A0Q9WTU0 B4LDR8 A0A2M4AGQ7 A0A0Q9WHL4 A0A2M3ZI29 A0A1B0CTD7 A0A0N0BJJ8 A0A034VIR4 A0A0K8URL1 A0A182FD32 A0A1I8MYF3 A0A034VH53 A0A0K8U8R6 A0A0K8VB69 W5JL99 A0A0L0CQQ8 A0A1J1ISV5 A0A1I8P764 A0A182WHQ3 A0A0N8P0U5 B3M5J1 A0A1A9WTF2 A0A3B0JT53 T1GHE2 A0A0M4ED41 B4KYD5 B4H633 A0A0Q9XP79 A0A1B6F5X8 A0A1B6EWE6 A0A3B0JT52 A0A3B0KN97 A0A0M4EB32 A0A3B0JT36 A0A3B0KHR6 B4JUE7
Pubmed
19121390
22118469
26354079
24845553
25401762
26823975
+ More
17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24438588 22936249 17510324 26999592 26483478 25244985 12364791 14747013 17210077 18362917 19820115 24495485 25830018 18057021 25348373 25315136 20920257 23761445 26108605
17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24438588 22936249 17510324 26999592 26483478 25244985 12364791 14747013 17210077 18362917 19820115 24495485 25830018 18057021 25348373 25315136 20920257 23761445 26108605
EMBL
BABH01024481
BABH01024482
ODYU01003219
SOQ41677.1
NWSH01000730
PCG74549.1
+ More
AGBW02012322 OWR45228.1 KQ461175 KPJ07850.1 KQ459302 KPJ01764.1 AGBW02011011 OWR47390.1 KK852824 KDR15450.1 GBHO01003544 GBRD01000085 JAG40060.1 JAG65736.1 GECU01013901 JAS93805.1 NEVH01013556 PNF28705.1 GDHC01008204 JAQ10425.1 CH933809 EDW17713.2 PNF28707.1 PNF28706.1 ACPB03026043 AE014296 AFH04338.1 AGB94273.1 CM000159 KRK01011.1 KRK01010.1 EDW93176.2 CCAG010023901 CCAG010023902 ATLV01026277 KE525409 KFB52975.1 CH954178 KQS43454.1 GBHO01014774 JAG28830.1 EDV50689.1 AXCM01001402 CM002912 KMY98375.1 KMY98377.1 KMY98376.1 KMY98378.1 CH478251 EAT33411.1 GDUN01000496 JAN95423.1 KK857864 PTY27522.1 JXUM01018208 KQ560505 KXJ82091.1 GEDC01027489 JAS09809.1 DS231814 EDS31755.1 AAF50426.2 AFH04339.1 AAAB01008960 EAA11687.4 EDO63785.1 EDO63786.1 AXCN02000398 UFQT01000016 SSX17836.1 KQ971312 KYB29146.1 GGFK01007765 MBW41086.1 EDW17715.1 GEBQ01017699 JAT22278.1 GAMC01011279 JAB95276.1 GDHF01031988 JAI20326.1 GAMC01011280 GAMC01011278 JAB95277.1 GBXI01013690 GBXI01007646 JAD00602.1 JAD06646.1 CH922616 EDW04567.1 CH916366 EDV96705.1 EDV96704.1 CH940647 EDW68940.1 KRF84091.1 EDW68941.2 GGFK01006645 MBW39966.1 KRF84087.1 KRF84088.1 KRF84089.1 GGFM01007418 MBW28169.1 AJWK01027435 AJWK01027436 KQ435713 KOX79261.1 GAKP01017506 GAKP01017504 JAC41448.1 GDHF01028832 GDHF01022990 GDHF01014239 GDHF01008989 JAI23482.1 JAI29324.1 JAI38075.1 JAI43325.1 GAKP01017505 JAC41447.1 GDHF01029331 JAI22983.1 GDHF01016574 JAI35740.1 ADMH02001237 ETN63544.1 JRES01000036 KNC34710.1 CVRI01000059 CRL03323.1 CH902618 KPU78099.1 KPU78100.1 EDV39601.1 OUUW01000009 SPP85285.1 CAQQ02033124 CP012525 ALC43446.1 EDW17714.2 CH479212 EDW33250.1 KRG05713.1 KRG05714.1 KRG05715.1 GECZ01024184 JAS45585.1 GECZ01027435 JAS42334.1 SPP85287.1 SPP85288.1 ALC44578.1 SPP85284.1 SPP85286.1 CH916374 EDV91117.1
AGBW02012322 OWR45228.1 KQ461175 KPJ07850.1 KQ459302 KPJ01764.1 AGBW02011011 OWR47390.1 KK852824 KDR15450.1 GBHO01003544 GBRD01000085 JAG40060.1 JAG65736.1 GECU01013901 JAS93805.1 NEVH01013556 PNF28705.1 GDHC01008204 JAQ10425.1 CH933809 EDW17713.2 PNF28707.1 PNF28706.1 ACPB03026043 AE014296 AFH04338.1 AGB94273.1 CM000159 KRK01011.1 KRK01010.1 EDW93176.2 CCAG010023901 CCAG010023902 ATLV01026277 KE525409 KFB52975.1 CH954178 KQS43454.1 GBHO01014774 JAG28830.1 EDV50689.1 AXCM01001402 CM002912 KMY98375.1 KMY98377.1 KMY98376.1 KMY98378.1 CH478251 EAT33411.1 GDUN01000496 JAN95423.1 KK857864 PTY27522.1 JXUM01018208 KQ560505 KXJ82091.1 GEDC01027489 JAS09809.1 DS231814 EDS31755.1 AAF50426.2 AFH04339.1 AAAB01008960 EAA11687.4 EDO63785.1 EDO63786.1 AXCN02000398 UFQT01000016 SSX17836.1 KQ971312 KYB29146.1 GGFK01007765 MBW41086.1 EDW17715.1 GEBQ01017699 JAT22278.1 GAMC01011279 JAB95276.1 GDHF01031988 JAI20326.1 GAMC01011280 GAMC01011278 JAB95277.1 GBXI01013690 GBXI01007646 JAD00602.1 JAD06646.1 CH922616 EDW04567.1 CH916366 EDV96705.1 EDV96704.1 CH940647 EDW68940.1 KRF84091.1 EDW68941.2 GGFK01006645 MBW39966.1 KRF84087.1 KRF84088.1 KRF84089.1 GGFM01007418 MBW28169.1 AJWK01027435 AJWK01027436 KQ435713 KOX79261.1 GAKP01017506 GAKP01017504 JAC41448.1 GDHF01028832 GDHF01022990 GDHF01014239 GDHF01008989 JAI23482.1 JAI29324.1 JAI38075.1 JAI43325.1 GAKP01017505 JAC41447.1 GDHF01029331 JAI22983.1 GDHF01016574 JAI35740.1 ADMH02001237 ETN63544.1 JRES01000036 KNC34710.1 CVRI01000059 CRL03323.1 CH902618 KPU78099.1 KPU78100.1 EDV39601.1 OUUW01000009 SPP85285.1 CAQQ02033124 CP012525 ALC43446.1 EDW17714.2 CH479212 EDW33250.1 KRG05713.1 KRG05714.1 KRG05715.1 GECZ01024184 JAS45585.1 GECZ01027435 JAS42334.1 SPP85287.1 SPP85288.1 ALC44578.1 SPP85284.1 SPP85286.1 CH916374 EDV91117.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000027135
+ More
UP000235965 UP000009192 UP000015103 UP000000803 UP000002282 UP000092444 UP000030765 UP000008711 UP000075883 UP000075880 UP000075885 UP000008820 UP000069940 UP000249989 UP000076408 UP000002320 UP000007062 UP000075881 UP000075903 UP000075886 UP000007266 UP000075900 UP000075884 UP000192221 UP000078200 UP000001070 UP000008792 UP000092461 UP000053105 UP000069272 UP000095301 UP000000673 UP000037069 UP000183832 UP000095300 UP000075920 UP000007801 UP000091820 UP000268350 UP000015102 UP000092553 UP000008744
UP000235965 UP000009192 UP000015103 UP000000803 UP000002282 UP000092444 UP000030765 UP000008711 UP000075883 UP000075880 UP000075885 UP000008820 UP000069940 UP000249989 UP000076408 UP000002320 UP000007062 UP000075881 UP000075903 UP000075886 UP000007266 UP000075900 UP000075884 UP000192221 UP000078200 UP000001070 UP000008792 UP000092461 UP000053105 UP000069272 UP000095301 UP000000673 UP000037069 UP000183832 UP000095300 UP000075920 UP000007801 UP000091820 UP000268350 UP000015102 UP000092553 UP000008744
Interpro
Gene 3D
ProteinModelPortal
H9JBL3
A0A2H1VLI2
A0A2A4JRV5
A0A212EUQ7
A0A194QS91
A0A194Q8N7
+ More
A0A212F0X9 A0A067R7H4 A0A0A9ZE12 A0A1B6J3R1 A0A2J7QJD5 A0A146LTH0 B4KYD4 A0A2J7QJF3 A0A2J7QJG0 T1HQF2 M9PEJ0 A0A0R1DYK0 A0A0R1E097 B4PG32 A0A1B0FM17 A0A084WRY1 A0A0Q5U387 A0A0A9YH28 B3NFS7 A0A182MPD0 A0A0J9UH40 A0A0J9RRE0 A0A182J4D0 A0A182P0X0 Q16GN7 A0A0P6IX53 A0A2R7X6X6 A0A182GTW9 A0A182Y378 A0A1B6C907 A0A1S4G1C9 B0VZL8 Q9VSJ3 Q7Q631 A0A182JS97 A0A182VP48 A7UU09 A0A182Q8B6 A0A336LIZ0 A0A139WMR3 A0A182RXH5 A0A2M4AJW8 B4KYD6 A0A182NKH8 A0A1W4US11 A0A1W4UF61 A0A1W4UEM4 A0A1B6LF10 W8BP58 A0A0K8U1F0 W8B9F7 A0A0A1X6X0 A0A1A9VYN0 B4K3H9 B4IZF0 B4IZE9 B4LDR7 A0A0Q9WTU0 B4LDR8 A0A2M4AGQ7 A0A0Q9WHL4 A0A2M3ZI29 A0A1B0CTD7 A0A0N0BJJ8 A0A034VIR4 A0A0K8URL1 A0A182FD32 A0A1I8MYF3 A0A034VH53 A0A0K8U8R6 A0A0K8VB69 W5JL99 A0A0L0CQQ8 A0A1J1ISV5 A0A1I8P764 A0A182WHQ3 A0A0N8P0U5 B3M5J1 A0A1A9WTF2 A0A3B0JT53 T1GHE2 A0A0M4ED41 B4KYD5 B4H633 A0A0Q9XP79 A0A1B6F5X8 A0A1B6EWE6 A0A3B0JT52 A0A3B0KN97 A0A0M4EB32 A0A3B0JT36 A0A3B0KHR6 B4JUE7
A0A212F0X9 A0A067R7H4 A0A0A9ZE12 A0A1B6J3R1 A0A2J7QJD5 A0A146LTH0 B4KYD4 A0A2J7QJF3 A0A2J7QJG0 T1HQF2 M9PEJ0 A0A0R1DYK0 A0A0R1E097 B4PG32 A0A1B0FM17 A0A084WRY1 A0A0Q5U387 A0A0A9YH28 B3NFS7 A0A182MPD0 A0A0J9UH40 A0A0J9RRE0 A0A182J4D0 A0A182P0X0 Q16GN7 A0A0P6IX53 A0A2R7X6X6 A0A182GTW9 A0A182Y378 A0A1B6C907 A0A1S4G1C9 B0VZL8 Q9VSJ3 Q7Q631 A0A182JS97 A0A182VP48 A7UU09 A0A182Q8B6 A0A336LIZ0 A0A139WMR3 A0A182RXH5 A0A2M4AJW8 B4KYD6 A0A182NKH8 A0A1W4US11 A0A1W4UF61 A0A1W4UEM4 A0A1B6LF10 W8BP58 A0A0K8U1F0 W8B9F7 A0A0A1X6X0 A0A1A9VYN0 B4K3H9 B4IZF0 B4IZE9 B4LDR7 A0A0Q9WTU0 B4LDR8 A0A2M4AGQ7 A0A0Q9WHL4 A0A2M3ZI29 A0A1B0CTD7 A0A0N0BJJ8 A0A034VIR4 A0A0K8URL1 A0A182FD32 A0A1I8MYF3 A0A034VH53 A0A0K8U8R6 A0A0K8VB69 W5JL99 A0A0L0CQQ8 A0A1J1ISV5 A0A1I8P764 A0A182WHQ3 A0A0N8P0U5 B3M5J1 A0A1A9WTF2 A0A3B0JT53 T1GHE2 A0A0M4ED41 B4KYD5 B4H633 A0A0Q9XP79 A0A1B6F5X8 A0A1B6EWE6 A0A3B0JT52 A0A3B0KN97 A0A0M4EB32 A0A3B0JT36 A0A3B0KHR6 B4JUE7
Ontologies
GO
PANTHER
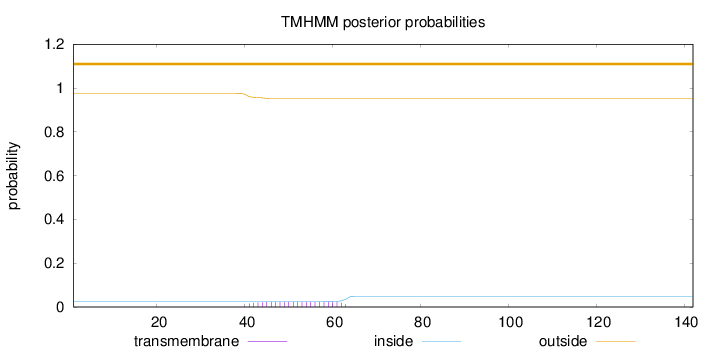
Topology
Length:
142
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.5173
Exp number, first 60 AAs:
0.46255
Total prob of N-in:
0.02387
outside
1 - 142
Population Genetic Test Statistics
Pi
237.627331
Theta
209.399554
Tajima's D
0.16337
CLR
0.101845
CSRT
0.412229388530573
Interpretation
Uncertain
