Gene
KWMTBOMO05816
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_sugar_transporter_SWEET1_[Bombyx_mori]
Full name
Sugar transporter SWEET
+ More
Sugar transporter SWEET1
Sugar transporter SWEET1
Alternative Name
Protein saliva
Location in the cell
PlasmaMembrane Reliability : 4.983
Sequence
CDS
ATGCATCTTTCTCAAATAAAGGATTTTGTTAGCAGTGTTGCTGTAACCATAACCATTTTACAGTTTCTTTCTGGAATTCTAGTTTGCAAACAATATGTCGTAAATCGGACCACAGCTGAAGCTTCTCCGCTTCCATTTATATGTGGCGTCATGTCTGCCGCTCTCTGGATATTGTATGGACTTACTAAACATGACAATAAAATTGTGTTTGTCAATGTAATTGGTGTGACGCTCATGTTTGCCTATACAGCTGTGTTCTACATCTTCACATTTAAAAAGTCATCAGTTTTGAAACAGATTTTTGCTATGGTTGCATTTATAATATTCACACTTGGATATGTGTCTGTTGAAGATGACAATGAACTACTTTTGGGTAGACTAGGGTTGTTAGCATGCTCCTTCACACTGTTGACGATAGCTTCTCCATTGAGTAAATTATTTTATGTCCTAAAAGTAAAATGTACTGAATGTCTTCCATTCCCAATGATACTAATGTCTTTTTTCGTGTCTGCAACATGGTTCCTATATGGCTTAATTGAAGAGGATGTATATATTACAACACCAAACTTCATTGGAGGTACATTGGCTGTACTTCAGCTCTCATTATTTTTAATCTTCCCGAGCAAACCCCATGGCTTGACAAAAAGTTTACTTGTTTTAAAAATAATTTTATTGACATTGTTATATTATTAA
Protein
MHLSQIKDFVSSVAVTITILQFLSGILVCKQYVVNRTTAEASPLPFICGVMSAALWILYGLTKHDNKIVFVNVIGVTLMFAYTAVFYIFTFKKSSVLKQIFAMVAFIIFTLGYVSVEDDNELLLGRLGLLACSFTLLTIASPLSKLFYVLKVKCTECLPFPMILMSFFVSATWFLYGLIEEDVYITTPNFIGGTLAVLQLSLFLIFPSKPHGLTKSLLVLKIILLTLLYY
Summary
Description
Mediates sugar transport across membranes.
Mediates both low-affinity uptake and efflux of sugar across the membrane.
Mediates both low-affinity uptake and efflux of sugar across the membrane.
Similarity
Belongs to the SWEET sugar transporter family.
Belongs to the E2F/DP family.
Belongs to the E2F/DP family.
Keywords
Cell membrane
Complete proteome
Golgi apparatus
Membrane
Reference proteome
Repeat
Sugar transport
Transmembrane
Transmembrane helix
Transport
Feature
chain Sugar transporter SWEET1
Uniprot
A0A2A4JT27
A0A212EUU2
A0A194QR77
A0A194Q9U5
A0A0N0PAD2
A0A2H1VLJ9
+ More
A0A0L7L795 A0A232FNU3 D6WK15 A0A1B0FV61 A0A1B0CYW3 A0A1L8DEV2 A0A0K8TS03 A0A023EKI3 A0A0A1XRW5 A0A1B6C0B4 Q176S9 A0A336LQ62 J3JYD4 A0A1Q3G2F9 T1HM70 A0A1I8MZZ3 A0A0A8JAH4 A0A0M5J555 A0A0V0G7F3 A0A1B0G5C1 A0A0P4VUX4 B4P283 A0A023EZ43 B4NMK1 A0A1A9YE65 A0A3B0J5Q6 B4J836 A0A0K8U1L7 U4U0V0 A0A0L0CLZ0 U5EYM8 E2BNB8 A0A1B6HCN6 A0A1A9ZCA0 A0A182P8V0 A0A0T6AYS0 A0A1A9VQJ6 E2A0T7 A0A1A9WWL0 B4GB34 E9JBP6 A0A182QKK1 A0A1W4WBI7 B4HRI6 B4QF97 A0A0B4LET7 Q7JVE7 Q7Q9H3 B3N994 A0A182IJH2 A0A182XET1 A0A182UUB1 A0A182HNV8 A0A158NHL9 Q290X1 A0A087ZY54 N6UNU8 A0A1I8PCN7 A0A2A3E5N5 A0A182WME3 A0A182MJ17 A0A2M3Z2Y6 B4LJL5 A0A182RV07 W5JNG5 A0A2M4BYL0 A0A2P8ZJT7 A0A2M3Z367 A0A1L8ECR6 A0A2M4ACK9 A0A1L8ECW9 A0A2M4ACB7 B4KMT6 A0A0A9Y973 A0A1W0WJV2 A0A0N8BC92 A0A0P5X4P1 A0A0L7R0L9 K7JEY1 A0A182FCV7 B3MGD6 A0A1B6H528 A0A1B6EBF2 A0A0M8ZXE6 A0A087UQQ6 A0A154PHQ7 A0A182U7L5 A0A1D1W9A2 E9FY84 A0A023FJE9 A0A151WGI2 A0A026W0N3 A0A195C1J5
A0A0L7L795 A0A232FNU3 D6WK15 A0A1B0FV61 A0A1B0CYW3 A0A1L8DEV2 A0A0K8TS03 A0A023EKI3 A0A0A1XRW5 A0A1B6C0B4 Q176S9 A0A336LQ62 J3JYD4 A0A1Q3G2F9 T1HM70 A0A1I8MZZ3 A0A0A8JAH4 A0A0M5J555 A0A0V0G7F3 A0A1B0G5C1 A0A0P4VUX4 B4P283 A0A023EZ43 B4NMK1 A0A1A9YE65 A0A3B0J5Q6 B4J836 A0A0K8U1L7 U4U0V0 A0A0L0CLZ0 U5EYM8 E2BNB8 A0A1B6HCN6 A0A1A9ZCA0 A0A182P8V0 A0A0T6AYS0 A0A1A9VQJ6 E2A0T7 A0A1A9WWL0 B4GB34 E9JBP6 A0A182QKK1 A0A1W4WBI7 B4HRI6 B4QF97 A0A0B4LET7 Q7JVE7 Q7Q9H3 B3N994 A0A182IJH2 A0A182XET1 A0A182UUB1 A0A182HNV8 A0A158NHL9 Q290X1 A0A087ZY54 N6UNU8 A0A1I8PCN7 A0A2A3E5N5 A0A182WME3 A0A182MJ17 A0A2M3Z2Y6 B4LJL5 A0A182RV07 W5JNG5 A0A2M4BYL0 A0A2P8ZJT7 A0A2M3Z367 A0A1L8ECR6 A0A2M4ACK9 A0A1L8ECW9 A0A2M4ACB7 B4KMT6 A0A0A9Y973 A0A1W0WJV2 A0A0N8BC92 A0A0P5X4P1 A0A0L7R0L9 K7JEY1 A0A182FCV7 B3MGD6 A0A1B6H528 A0A1B6EBF2 A0A0M8ZXE6 A0A087UQQ6 A0A154PHQ7 A0A182U7L5 A0A1D1W9A2 E9FY84 A0A023FJE9 A0A151WGI2 A0A026W0N3 A0A195C1J5
Pubmed
22118469
26354079
26227816
28648823
18362917
19820115
+ More
26369729 24945155 25830018 17510324 22516182 25315136 27129103 17994087 17550304 25474469 23537049 26108605 20798317 21282665 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9739134 12537569 12364791 14747013 17210077 21347285 15632085 18057021 20920257 23761445 29403074 25401762 26823975 20075255 27649274 21292972 24508170
26369729 24945155 25830018 17510324 22516182 25315136 27129103 17994087 17550304 25474469 23537049 26108605 20798317 21282665 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9739134 12537569 12364791 14747013 17210077 21347285 15632085 18057021 20920257 23761445 29403074 25401762 26823975 20075255 27649274 21292972 24508170
EMBL
NWSH01000730
PCG74550.1
AGBW02012322
OWR45227.1
KQ461175
KPJ07849.1
+ More
KQ459302 KPJ01765.1 KQ458960 KPJ04540.1 ODYU01003219 SOQ41676.1 JTDY01002543 KOB71214.1 NNAY01000006 OXU32128.1 KQ971342 EFA04753.1 AJWK01010391 AJVK01009378 GFDF01009108 JAV04976.1 GDAI01000898 JAI16705.1 GAPW01003943 JAC09655.1 GBXI01001024 JAD13268.1 GEDC01030347 GEDC01029656 JAS06951.1 JAS07642.1 CH477383 EAT42162.1 UFQT01000040 SSX18537.1 BT128260 AEE63220.1 GFDL01001061 JAV33984.1 ACPB03017303 AB901081 BAQ02381.1 CP012524 ALC42194.1 GECL01002484 JAP03640.1 CCAG010011642 GDKW01001980 JAI54615.1 CM000157 EDW89284.1 GBBI01004229 JAC14483.1 CH964282 EDW85590.1 OUUW01000001 SPP75153.1 CH916367 EDW02266.1 GDHF01031913 JAI20401.1 KB630546 ERL83670.1 JRES01000204 KNC33315.1 GANO01001908 JAB57963.1 GL449385 EFN82880.1 GECU01035344 GECU01004755 JAS72362.1 JAT02952.1 LJIG01022500 KRT80241.1 GL435626 EFN72990.1 CH479181 EDW32136.1 GL771340 EFZ09763.1 AXCN02001017 CH480816 EDW46868.1 CM000362 CM002911 EDX06126.1 KMY92178.1 AE013599 AHN55980.1 U70979 AY118993 AAAB01008900 EAA09398.3 CH954177 EDV59581.1 APCN01002793 ADTU01015965 ADTU01015966 ADTU01015967 CM000071 APGK01003259 APGK01003260 KB735185 ENN83425.1 KZ288369 PBC26794.1 AXCM01002945 GGFM01002136 MBW22887.1 CH940648 EDW61583.1 KRF80088.1 ADMH02000552 ETN65897.1 GGFJ01009035 MBW58176.1 PYGN01000035 PSN56762.1 GGFM01002137 MBW22888.1 GFDG01002302 JAV16497.1 GGFK01005169 MBW38490.1 GFDG01002303 JAV16496.1 GGFK01005115 MBW38436.1 CH933808 EDW08828.1 GBHO01015978 GBHO01015977 GBHO01015976 GBHO01015975 GBRD01013412 GDHC01014283 GDHC01001207 JAG27626.1 JAG27627.1 JAG27628.1 JAG27629.1 JAG52414.1 JAQ04346.1 JAQ17422.1 MTYJ01000088 OQV15501.1 GDIQ01191923 GDIP01045824 JAK59802.1 JAM57891.1 GDIP01077907 JAM25808.1 KQ414670 KOC64402.1 AAZX01003330 CH902619 EDV37839.1 GECZ01000065 JAS69704.1 GEDC01002040 JAS35258.1 KQ435826 KOX72026.1 KK121079 KFM79695.1 KQ434912 KZC11379.1 BDGG01000015 GAV07669.1 GL732527 EFX87821.1 GBBK01003684 JAC20798.1 KQ983167 KYQ46934.1 KK107591 EZA48619.1 KQ978350 KYM94717.1
KQ459302 KPJ01765.1 KQ458960 KPJ04540.1 ODYU01003219 SOQ41676.1 JTDY01002543 KOB71214.1 NNAY01000006 OXU32128.1 KQ971342 EFA04753.1 AJWK01010391 AJVK01009378 GFDF01009108 JAV04976.1 GDAI01000898 JAI16705.1 GAPW01003943 JAC09655.1 GBXI01001024 JAD13268.1 GEDC01030347 GEDC01029656 JAS06951.1 JAS07642.1 CH477383 EAT42162.1 UFQT01000040 SSX18537.1 BT128260 AEE63220.1 GFDL01001061 JAV33984.1 ACPB03017303 AB901081 BAQ02381.1 CP012524 ALC42194.1 GECL01002484 JAP03640.1 CCAG010011642 GDKW01001980 JAI54615.1 CM000157 EDW89284.1 GBBI01004229 JAC14483.1 CH964282 EDW85590.1 OUUW01000001 SPP75153.1 CH916367 EDW02266.1 GDHF01031913 JAI20401.1 KB630546 ERL83670.1 JRES01000204 KNC33315.1 GANO01001908 JAB57963.1 GL449385 EFN82880.1 GECU01035344 GECU01004755 JAS72362.1 JAT02952.1 LJIG01022500 KRT80241.1 GL435626 EFN72990.1 CH479181 EDW32136.1 GL771340 EFZ09763.1 AXCN02001017 CH480816 EDW46868.1 CM000362 CM002911 EDX06126.1 KMY92178.1 AE013599 AHN55980.1 U70979 AY118993 AAAB01008900 EAA09398.3 CH954177 EDV59581.1 APCN01002793 ADTU01015965 ADTU01015966 ADTU01015967 CM000071 APGK01003259 APGK01003260 KB735185 ENN83425.1 KZ288369 PBC26794.1 AXCM01002945 GGFM01002136 MBW22887.1 CH940648 EDW61583.1 KRF80088.1 ADMH02000552 ETN65897.1 GGFJ01009035 MBW58176.1 PYGN01000035 PSN56762.1 GGFM01002137 MBW22888.1 GFDG01002302 JAV16497.1 GGFK01005169 MBW38490.1 GFDG01002303 JAV16496.1 GGFK01005115 MBW38436.1 CH933808 EDW08828.1 GBHO01015978 GBHO01015977 GBHO01015976 GBHO01015975 GBRD01013412 GDHC01014283 GDHC01001207 JAG27626.1 JAG27627.1 JAG27628.1 JAG27629.1 JAG52414.1 JAQ04346.1 JAQ17422.1 MTYJ01000088 OQV15501.1 GDIQ01191923 GDIP01045824 JAK59802.1 JAM57891.1 GDIP01077907 JAM25808.1 KQ414670 KOC64402.1 AAZX01003330 CH902619 EDV37839.1 GECZ01000065 JAS69704.1 GEDC01002040 JAS35258.1 KQ435826 KOX72026.1 KK121079 KFM79695.1 KQ434912 KZC11379.1 BDGG01000015 GAV07669.1 GL732527 EFX87821.1 GBBK01003684 JAC20798.1 KQ983167 KYQ46934.1 KK107591 EZA48619.1 KQ978350 KYM94717.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
UP000215335
+ More
UP000007266 UP000092461 UP000092462 UP000008820 UP000015103 UP000095301 UP000092553 UP000092444 UP000002282 UP000007798 UP000092443 UP000268350 UP000001070 UP000030742 UP000037069 UP000008237 UP000092445 UP000075885 UP000078200 UP000000311 UP000091820 UP000008744 UP000075886 UP000192221 UP000001292 UP000000304 UP000000803 UP000007062 UP000008711 UP000075880 UP000076407 UP000075903 UP000075840 UP000005205 UP000001819 UP000005203 UP000019118 UP000095300 UP000242457 UP000075920 UP000075883 UP000008792 UP000075900 UP000000673 UP000245037 UP000009192 UP000053825 UP000002358 UP000069272 UP000007801 UP000053105 UP000054359 UP000076502 UP000075902 UP000186922 UP000000305 UP000075809 UP000053097 UP000078542
UP000007266 UP000092461 UP000092462 UP000008820 UP000015103 UP000095301 UP000092553 UP000092444 UP000002282 UP000007798 UP000092443 UP000268350 UP000001070 UP000030742 UP000037069 UP000008237 UP000092445 UP000075885 UP000078200 UP000000311 UP000091820 UP000008744 UP000075886 UP000192221 UP000001292 UP000000304 UP000000803 UP000007062 UP000008711 UP000075880 UP000076407 UP000075903 UP000075840 UP000005205 UP000001819 UP000005203 UP000019118 UP000095300 UP000242457 UP000075920 UP000075883 UP000008792 UP000075900 UP000000673 UP000245037 UP000009192 UP000053825 UP000002358 UP000069272 UP000007801 UP000053105 UP000054359 UP000076502 UP000075902 UP000186922 UP000000305 UP000075809 UP000053097 UP000078542
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JT27
A0A212EUU2
A0A194QR77
A0A194Q9U5
A0A0N0PAD2
A0A2H1VLJ9
+ More
A0A0L7L795 A0A232FNU3 D6WK15 A0A1B0FV61 A0A1B0CYW3 A0A1L8DEV2 A0A0K8TS03 A0A023EKI3 A0A0A1XRW5 A0A1B6C0B4 Q176S9 A0A336LQ62 J3JYD4 A0A1Q3G2F9 T1HM70 A0A1I8MZZ3 A0A0A8JAH4 A0A0M5J555 A0A0V0G7F3 A0A1B0G5C1 A0A0P4VUX4 B4P283 A0A023EZ43 B4NMK1 A0A1A9YE65 A0A3B0J5Q6 B4J836 A0A0K8U1L7 U4U0V0 A0A0L0CLZ0 U5EYM8 E2BNB8 A0A1B6HCN6 A0A1A9ZCA0 A0A182P8V0 A0A0T6AYS0 A0A1A9VQJ6 E2A0T7 A0A1A9WWL0 B4GB34 E9JBP6 A0A182QKK1 A0A1W4WBI7 B4HRI6 B4QF97 A0A0B4LET7 Q7JVE7 Q7Q9H3 B3N994 A0A182IJH2 A0A182XET1 A0A182UUB1 A0A182HNV8 A0A158NHL9 Q290X1 A0A087ZY54 N6UNU8 A0A1I8PCN7 A0A2A3E5N5 A0A182WME3 A0A182MJ17 A0A2M3Z2Y6 B4LJL5 A0A182RV07 W5JNG5 A0A2M4BYL0 A0A2P8ZJT7 A0A2M3Z367 A0A1L8ECR6 A0A2M4ACK9 A0A1L8ECW9 A0A2M4ACB7 B4KMT6 A0A0A9Y973 A0A1W0WJV2 A0A0N8BC92 A0A0P5X4P1 A0A0L7R0L9 K7JEY1 A0A182FCV7 B3MGD6 A0A1B6H528 A0A1B6EBF2 A0A0M8ZXE6 A0A087UQQ6 A0A154PHQ7 A0A182U7L5 A0A1D1W9A2 E9FY84 A0A023FJE9 A0A151WGI2 A0A026W0N3 A0A195C1J5
A0A0L7L795 A0A232FNU3 D6WK15 A0A1B0FV61 A0A1B0CYW3 A0A1L8DEV2 A0A0K8TS03 A0A023EKI3 A0A0A1XRW5 A0A1B6C0B4 Q176S9 A0A336LQ62 J3JYD4 A0A1Q3G2F9 T1HM70 A0A1I8MZZ3 A0A0A8JAH4 A0A0M5J555 A0A0V0G7F3 A0A1B0G5C1 A0A0P4VUX4 B4P283 A0A023EZ43 B4NMK1 A0A1A9YE65 A0A3B0J5Q6 B4J836 A0A0K8U1L7 U4U0V0 A0A0L0CLZ0 U5EYM8 E2BNB8 A0A1B6HCN6 A0A1A9ZCA0 A0A182P8V0 A0A0T6AYS0 A0A1A9VQJ6 E2A0T7 A0A1A9WWL0 B4GB34 E9JBP6 A0A182QKK1 A0A1W4WBI7 B4HRI6 B4QF97 A0A0B4LET7 Q7JVE7 Q7Q9H3 B3N994 A0A182IJH2 A0A182XET1 A0A182UUB1 A0A182HNV8 A0A158NHL9 Q290X1 A0A087ZY54 N6UNU8 A0A1I8PCN7 A0A2A3E5N5 A0A182WME3 A0A182MJ17 A0A2M3Z2Y6 B4LJL5 A0A182RV07 W5JNG5 A0A2M4BYL0 A0A2P8ZJT7 A0A2M3Z367 A0A1L8ECR6 A0A2M4ACK9 A0A1L8ECW9 A0A2M4ACB7 B4KMT6 A0A0A9Y973 A0A1W0WJV2 A0A0N8BC92 A0A0P5X4P1 A0A0L7R0L9 K7JEY1 A0A182FCV7 B3MGD6 A0A1B6H528 A0A1B6EBF2 A0A0M8ZXE6 A0A087UQQ6 A0A154PHQ7 A0A182U7L5 A0A1D1W9A2 E9FY84 A0A023FJE9 A0A151WGI2 A0A026W0N3 A0A195C1J5
PDB
5XPD
E-value=2.81932e-11,
Score=162
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Cell membrane
Golgi apparatus membrane
Nucleus
Golgi apparatus membrane
Nucleus
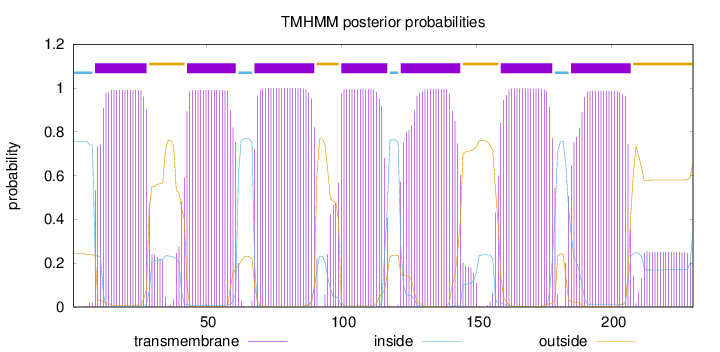
Length:
230
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
151.15136
Exp number, first 60 AAs:
39.45539
Total prob of N-in:
0.75653
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 28
outside
29 - 42
TMhelix
43 - 61
inside
62 - 67
TMhelix
68 - 90
outside
91 - 99
TMhelix
100 - 117
inside
118 - 121
TMhelix
122 - 144
outside
145 - 158
TMhelix
159 - 178
inside
179 - 184
TMhelix
185 - 207
outside
208 - 230
Population Genetic Test Statistics
Pi
29.141541
Theta
183.419358
Tajima's D
1.746392
CLR
0.298013
CSRT
0.84030798460077
Interpretation
Uncertain
