Gene
KWMTBOMO05813
Pre Gene Modal
BGIBMGA006909
Annotation
PREDICTED:_DEP_domain-containing_protein_5_[Bombyx_mori]
Full name
GATOR complex protein Iml1
Alternative Name
Increased minichromosome loss 1
Location in the cell
Nuclear Reliability : 2.127
Sequence
CDS
ATGAAGTCCTTTAAACTCATTGTACATCAGTCTAGTTTTAGCCCCGAAGATTTGATTATCAACTTGAAAGACTATCCAGGGTTAAAGGAAAAAGATATAGTCGAAATTTATCATCCCGAAAATGACTACCCCCGATTATTACTACAAGTTACAAAAGTGGACGCCCCCGGACGCGGCAGAGATACAATTAGTGTGGAACAAAGTATTGCTACCACTTTCCAACTCCGAACGTTTGCTGATGTCTATGTAAATATAGTCAATGTGCAAGATGTGGCTTTGGATTCAGTGGAATTAACATTCAAAGACCAATATCTCGGAAGATCCGAAATGTGGAGACTTAAAAATCACCTAGTAAGCACCTGCGTTTATTTAAACAAGAAGATAGAATATTGTGGTGGTGCAATTAGATGTCAAGTATATGAAATGTGGTCACAAGGGGACAGAGTTGCTTGTGGGGTAATCACTGAAGACACAAAAATAGTGTTCAGATCATCAACTTCAATGGTCTACCTCTTCATTCAAATGTCTTCTGAAATGTGGGATTTTGACATCCATGGTGATCTGTACTTTGAAAAGGCAGTTAATGGTTTCCTCGCTGATTTGTTTGCCAAATGGAAGAAAAATGGCAGTAATCATGAAGTAACCATAGTTCTCTTTTCAAGAACTTTTTATAAAGCTAAGTCTTTAGAAGAATTTCCTCAACATATGCAAGAGTGCTTGCAAAAAGATTACAGGGGGAGATTTTATGAAGACTTTTACAGGGTTGCAGTACAAAATGAGAGATATGAAGACTGGTCAAATGTACTACTACAATTAAGAAGACTCTTTACTGATTATCAAAAGATTGTGCTTCAGTACCATGAAAGGCCTAACATGGACATACCCACAGCAATAAACTCCACTGCTGCACAAGGAAATTTCCTAGAAGTCCTTAACATGAGTTTGAATGTATTCGAAAAACATTACCTGGATAGATGTTTCGACAGAACGGGGCAGCTGAGTGTGGTAATAACCCCAGGAGTAGGTGTATTTGAAGTCGACAGAGAACTGACTAATGTTACAAAACAAAGAATCATTGACAATGGTGTTGGCAGTGACTTAGTTTGTGTAGGAGAACAACCTCTGCATGCTGTGCCTTTACTAAAGTTCCACAATAAAGACAATAATTTGAATTCAATTGATGATTATTCGATGCCACATTGGATTAATCTATCATTTTATTCTACAAATAAAAAAGTGGCTTATTCCAATTTCATACCAAGAATTAAATTGCCTCCACGAAAGAGCCAAGAACCATTACGTAAAATGTATGAAGATGAGAGGAAAGGTAAATTGTTGAAAGAAGATGAATGTATGCACAATTCTATATTTGATTATGATGCATACGACGCGCAAGTTTTTCAGTTACCCCCCGCTCACAGCACACGGGTACAAAGGGTCGCCAGGACTAAGAAAACTTCTGTGGCGGGTTTAGAAGGCTTAAATCGTAGGAGCCCTCCAGCTTCTACTTTGCATCACAGAAAAATGTCGGATCCGGACATCCATCATAGCCTTGGAGATCTTTTGACTTCTGGAAACAAGTCCGCAGGCTACGACGCGGGCAGTGACACAACGGACTCGCCGATGTCGACGAACCGACTCTCGCCGCGCAGTTCGATATCTTCAATCAAGCCCGTTATCAGGACGGGCAGGGCACTCATCAACCCCTTCGATCCCTCGCATGTCACTGTCAAACTGACCAGTAACCGTCGAAGGTGGACGCATATATTTCCGAAAGGTCCAACTGGCGTATTGATACAGCAACATCACTACCAGGCCCGTCCCGCTGGCGAGCCGGCCCGCAACGAAGAGCATCGCTGCGAAAGTAACACACCAAAGGATAATGGTTCCTCGATATTAAACAACAACCATTCTATATATCAGGTTGACGGCAACATTATGTCGCGGAAACGCATGATAAGTGCGTCAGGAACGTTAGCCGTGCTCGGCGTGTCCGGGGGGACTCCCTCCACGACCAACAATGCGTCGCTCGCGCTGCTGTGGGGGGCTACGGGCGAACAGGAGTGGACCCCCGCTCTGACGACAGGGGTCGACTGGAAGTCACTGACCATACCTGCCTGCCTTCCGATCACTACGGACTACTTTCCCGACAAGAGATCACTTCAAAACGATTATCTAGTATCGGATTATAATCTGTTACCGGACGACGTGAACGCCGATTTCGCACAGAACCGAGCTATATACAAAGATCCCCTGACCACAATGGAAGTATTCAAGGAGTTGGTCTCACAAAGATTAGCTCAGGGTTTCCAGCTGATTGTGGGTGTTAACGAGAATGAAGTAATCGAATCTCACTGTGCGTCAACGGCGACTCCCCCACCATCCAAAGTGGCCCCACAGTGCGGGAAACCAGCCAACGCAGCACCCACCAAGCGTTACCTGCTCTCGATTGGGAGGATCTTCCACAAACTTACGCTGGTCGGGTCCACCATCACCGTCACTAGATACAGACCCAGACATCCATATCCGCCATTCAATATACACTACCGTTATCGTTTCCACGCACCGAACCACGACACGTACGAGGTGTCGTGGGTTTCGTTCACGACAGAAAAACTGGAAACTTACTACTGGAACTACATGGACCATTACATTTGTACGCGCGGCGACAGTGATTTCGCACTAGTTGAGCCTCTGAAGTACTGGCGGTTCCGGACTCTACTATTGCCTCTCTACAATCCGGCAACGAAACAAATTCTCGAAGACGATTCGACCCACTGCGACATATATCCAACTCCGACGCGTCATGATTTGGACCAACTAACCGATAATTTCCTCAAAATGACCGAATTGTATTTCAATAAAGTCAAAAGGCCAAACAAACAGAGGATGGCGGGCGCGGGCTGCGGCGCTAGCGACGCGGCACTCACGCGGCGCCGACACTCCACCTCCATCCTCACGCGGAGCGCACAGGCAGGGGTATCGAGCTCTCCATTCAGAGAGCGCGTTGGCAGCACCCGGCTACCTGACCGGCCTCGCCTCCGGATGGAAGCTATGGCGACGAAAGCAGTGCTAGAAAGGACGCAATCCCAAACCGGAGAAGCGGATGACACTTATGATGACGGAGTTTCAGAACCGAAACTGAAAGCGAACGCGCCATTGTCGGATGTCGTAGATAGAATGCGACACCCGACTCTGGGCTTGGGTTTCCTACAACAGACGGTCAGTTTGCCCTCGCATACATTCGTTTCGATATACGCCATACATTGGCTCCAAGAAAACATGGAGGGAATGGATTATGAAAAGGCTGCCAGTCTTATGGAGAGATTATTGAATGAGAAAATGATTTGTCATGCTTCAGGCGACACCACAAAACGTTTCGTTGTCGGATACTACATGTATCACATACTACCACAAAAGAAAGATAAAGAATTACCAGACTACGTGAAGCCCCTAGGAGACCTCCAGAGCTTCGAAAACGAATGGATGGAAGTTGAGGTACTGGGGCCGCGTTCGCCGCTCATACAAGTGGACAAAGCGAGCGGCGCCATAGATATATCTGGTTCTAGTCCTCTGGCCGACGACACGGGCGTTCCCGCCTTCCTGTGCGATAATATTGATCCAAACTACATGCAAATCGATAGCGACAGCGAAATACCCTTGTACAAGAAAACCCACCTAGATATAGATGTGAATAATAAAAGCGACCGGATCGAATGGGGCCACGCACGATATCAAGCCACGTTCCGACCGGACCAAGCCTTCGAAATGTGCATACAGTGGACAGTCGCCAGCGGAAATATTGTTGCCGAACTGATATTCGGATGGGCTCGTAAAGCCTCGTCCAACTGTAAGCTACAAATGGTGCCTATTCCTGCGGACCCCCTCGCTTTACCATTCACCCTCAAATCAGACCCGTTGCGAGGTCCGATTTACATTCCTATGAACGTAGAACCATTGTTGAGAAATAAGACTGTTTTGTTTGAAGGCTTCCCGGAAGAGACATGGCTCCAGCGTTTGTTCCTATTCCAAGAGGCCATAGTGGGTCGTTTCGGCTTCGTGAAGTGCACGGTCGAGAGCACGTCGCACGAGTGGGAGCGCGGCACGCCCCGCGGCTCGCCCACCGGGGACCACCTGTACGTGCACGTGACCGGGAACATGTTCATACTCATACCGACGACGGTCAAGTCCGAGCAGAAGTCGTCGAAGGTCAAGCCGCCGAACAGAACGCTGTGTAACAAGTACCCTGTGCCGACAGAAGTTGCCAACAGTCCTCATGAAGCTTATATTTCTCGACATGTCGGGTACAAAACGAACGACTACAATAATAGCAGACGGATGGGATTTTTATGGTCTTGGAATCACATGATTTCAAAGAAATGGAAGGGTTCACAAACACCAGCCACAGGAGACGAAGGTTTCCAAATGCGAATGCTAAGGGATTTCAAACATTTCTGTGCCAATCAGGAACAGAGGCTTAGTCAATTTTGGGATCACTGCTGGGAACTAAAGGATAAATCCCGAGGAATGACATCTTGTTAA
Protein
MKSFKLIVHQSSFSPEDLIINLKDYPGLKEKDIVEIYHPENDYPRLLLQVTKVDAPGRGRDTISVEQSIATTFQLRTFADVYVNIVNVQDVALDSVELTFKDQYLGRSEMWRLKNHLVSTCVYLNKKIEYCGGAIRCQVYEMWSQGDRVACGVITEDTKIVFRSSTSMVYLFIQMSSEMWDFDIHGDLYFEKAVNGFLADLFAKWKKNGSNHEVTIVLFSRTFYKAKSLEEFPQHMQECLQKDYRGRFYEDFYRVAVQNERYEDWSNVLLQLRRLFTDYQKIVLQYHERPNMDIPTAINSTAAQGNFLEVLNMSLNVFEKHYLDRCFDRTGQLSVVITPGVGVFEVDRELTNVTKQRIIDNGVGSDLVCVGEQPLHAVPLLKFHNKDNNLNSIDDYSMPHWINLSFYSTNKKVAYSNFIPRIKLPPRKSQEPLRKMYEDERKGKLLKEDECMHNSIFDYDAYDAQVFQLPPAHSTRVQRVARTKKTSVAGLEGLNRRSPPASTLHHRKMSDPDIHHSLGDLLTSGNKSAGYDAGSDTTDSPMSTNRLSPRSSISSIKPVIRTGRALINPFDPSHVTVKLTSNRRRWTHIFPKGPTGVLIQQHHYQARPAGEPARNEEHRCESNTPKDNGSSILNNNHSIYQVDGNIMSRKRMISASGTLAVLGVSGGTPSTTNNASLALLWGATGEQEWTPALTTGVDWKSLTIPACLPITTDYFPDKRSLQNDYLVSDYNLLPDDVNADFAQNRAIYKDPLTTMEVFKELVSQRLAQGFQLIVGVNENEVIESHCASTATPPPSKVAPQCGKPANAAPTKRYLLSIGRIFHKLTLVGSTITVTRYRPRHPYPPFNIHYRYRFHAPNHDTYEVSWVSFTTEKLETYYWNYMDHYICTRGDSDFALVEPLKYWRFRTLLLPLYNPATKQILEDDSTHCDIYPTPTRHDLDQLTDNFLKMTELYFNKVKRPNKQRMAGAGCGASDAALTRRRHSTSILTRSAQAGVSSSPFRERVGSTRLPDRPRLRMEAMATKAVLERTQSQTGEADDTYDDGVSEPKLKANAPLSDVVDRMRHPTLGLGFLQQTVSLPSHTFVSIYAIHWLQENMEGMDYEKAASLMERLLNEKMICHASGDTTKRFVVGYYMYHILPQKKDKELPDYVKPLGDLQSFENEWMEVEVLGPRSPLIQVDKASGAIDISGSSPLADDTGVPAFLCDNIDPNYMQIDSDSEIPLYKKTHLDIDVNNKSDRIEWGHARYQATFRPDQAFEMCIQWTVASGNIVAELIFGWARKASSNCKLQMVPIPADPLALPFTLKSDPLRGPIYIPMNVEPLLRNKTVLFEGFPEETWLQRLFLFQEAIVGRFGFVKCTVESTSHEWERGTPRGSPTGDHLYVHVTGNMFILIPTTVKSEQKSSKVKPPNRTLCNKYPVPTEVANSPHEAYISRHVGYKTNDYNNSRRMGFLWSWNHMISKKWKGSQTPATGDEGFQMRMLRDFKHFCANQEQRLSQFWDHCWELKDKSRGMTSC
Summary
Description
An essential component of the GATOR subcomplex GATOR1 which functions as an inhibitor of the amino acid-sensing branch of the TORC1 signaling pathway (PubMed:23723238, PubMed:27672113, PubMed:25512509). The two GATOR subcomplexes, GATOR1 and GATOR2, regulate the TORC1 pathway in order to mediate metabolic homeostasis, female gametogenesis and the response to amino acid limitation and complete starvation (PubMed:23723238, PubMed:27672113, PubMed:25512509). The function of GATOR1 in negatively regulating the TORC1 pathway is essential for maintaining baseline levels of TORC1 activity under nutrient rich conditions, and for promoting survival during amino acid or complete starvation by inhibiting TORC1-dependent cell growth and promoting catabolic metabolism and autophagy (PubMed:23723238, PubMed:27672113, PubMed:25512509). GATOR1 and GATOR2 act at different stages of oogenesis to regulate TORC1 in order to control meiotic entry and promote oocyte growth and development (PubMed:27672113, PubMed:25512509). After exactly four mitotic cyst divisions, the GATOR1 complex members (Iml1, Nprl2 and Nprl3) down-regulate TORC1 to slow cellular metabolism and promote the mitotic/meiotic transition (PubMed:27672113, PubMed:25512509). At later stages of oogenesis, the mio and Nup44A components of the GATOR2 complex inhibit GATOR1 and thus activate TORC1 to promote meiotic progression, and drive oocyte growth and development (PubMed:27672113, PubMed:25512509).
Subunit
Component of the GATOR complex consisting of mio, Nup44A/Seh1, Im11, Nplr3, Nplr2, Wdr24, Wdr59 and Sec13 (PubMed:27166823). Within the GATOR complex, probable component of the GATOR1 subcomplex which is likely composed of Iml1, Nplr2 and Nplr3 (PubMed:27166823).
Similarity
Belongs to the IML1 family.
Keywords
Alternative splicing
Cell cycle
Cell division
Complete proteome
Meiosis
Mitosis
Reference proteome
Feature
chain GATOR complex protein Iml1
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A2A4JSW6
A0A2A4JR85
A0A2A4JT36
A0A2A4JSG8
A0A3S2P8B4
A0A2A4JRD6
+ More
A0A1E1WB64 A0A212EUR6 A0A1E1W9K7 A0A194QQR6 A0A194QE74 H9JBL4 E2BB04 A0A158P0Q8 A0A0L7R3R3 A0A2A3EIL1 A0A0M8ZTI0 A0A088A8W3 A0A3L8E1I9 K7IT54 E2AFB4 A0A232FL67 A0A154PG92 A0A151WWI1 E0VTF7 A0A1Y1KEN3 A0A1Y1KCF9 A0A1L8DMQ9 A0A195FX74 A0A1Q3FE62 A0A026WDB7 A0A023F2J6 A0A1S4FXU9 A0A1Q3FE70 A0A1B6DCJ2 B0WMQ7 Q16KP8 D6X293 A0A182FDQ0 A0A182WL50 A0A2M4ABU9 N6U9Z9 A0A182UW39 A0A0A1X6Z1 W8AWB3 A0A1I8MH41 Q7PF61 A0A146M7J6 A0A1B0FMG6 A0A1B6FI79 A0A1I8MH47 A0A1Y1KCE5 W8B3W3 A0A0L0CL22 A0A1I8MH44 A0A1A9ZU36 A0A1A9V409 A0A1Y1KBU7 A0A1J1IT55 A0A1B6C5V5 A0A1I8MH54 A0A1I8PYI3 A0A1B6H3H8 A0A1I8MH43 A0A1I8MH46 A0A1J1IUP4 A0A0Q5UGY3 A0A0Q5UH86 A0A1I8MH49 A0A0A1X7M9 A0A1B0GJN7 A0A1B6F1V2 Q29FE7 A0A0Q9XR66 A0A0Q9WHT5 M9PDV3 A0A0R1DUA6 A0A1W4W610 B3NBB8 A0A0J9RLN3 A0A0Q5UGT4 A0A0Q9XT29 A0A0R3P2I6 A0A0R3P7M6 A0A0Q9WGV2 A0A0R1DUJ8 A0A3B0JUX3 B4PD98 A0A0N8P0P6 B4MKU6 Q9W0E3 Q8IRG8 A0A0P9AIP1 A0A1W4W6K3 A0A0J9UC37 A0A1W4VTZ7 A0A0J9RN56 B4L8V3 A0A0Q5U2Y5
A0A1E1WB64 A0A212EUR6 A0A1E1W9K7 A0A194QQR6 A0A194QE74 H9JBL4 E2BB04 A0A158P0Q8 A0A0L7R3R3 A0A2A3EIL1 A0A0M8ZTI0 A0A088A8W3 A0A3L8E1I9 K7IT54 E2AFB4 A0A232FL67 A0A154PG92 A0A151WWI1 E0VTF7 A0A1Y1KEN3 A0A1Y1KCF9 A0A1L8DMQ9 A0A195FX74 A0A1Q3FE62 A0A026WDB7 A0A023F2J6 A0A1S4FXU9 A0A1Q3FE70 A0A1B6DCJ2 B0WMQ7 Q16KP8 D6X293 A0A182FDQ0 A0A182WL50 A0A2M4ABU9 N6U9Z9 A0A182UW39 A0A0A1X6Z1 W8AWB3 A0A1I8MH41 Q7PF61 A0A146M7J6 A0A1B0FMG6 A0A1B6FI79 A0A1I8MH47 A0A1Y1KCE5 W8B3W3 A0A0L0CL22 A0A1I8MH44 A0A1A9ZU36 A0A1A9V409 A0A1Y1KBU7 A0A1J1IT55 A0A1B6C5V5 A0A1I8MH54 A0A1I8PYI3 A0A1B6H3H8 A0A1I8MH43 A0A1I8MH46 A0A1J1IUP4 A0A0Q5UGY3 A0A0Q5UH86 A0A1I8MH49 A0A0A1X7M9 A0A1B0GJN7 A0A1B6F1V2 Q29FE7 A0A0Q9XR66 A0A0Q9WHT5 M9PDV3 A0A0R1DUA6 A0A1W4W610 B3NBB8 A0A0J9RLN3 A0A0Q5UGT4 A0A0Q9XT29 A0A0R3P2I6 A0A0R3P7M6 A0A0Q9WGV2 A0A0R1DUJ8 A0A3B0JUX3 B4PD98 A0A0N8P0P6 B4MKU6 Q9W0E3 Q8IRG8 A0A0P9AIP1 A0A1W4W6K3 A0A0J9UC37 A0A1W4VTZ7 A0A0J9RN56 B4L8V3 A0A0Q5U2Y5
Pubmed
22118469
26354079
19121390
20798317
21347285
30249741
+ More
20075255 28648823 20566863 28004739 24508170 25474469 17510324 18362917 19820115 23537049 25830018 24495485 25315136 12364791 14747013 17210077 26823975 26108605 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 12537569 23723238 25512509 27672113 27166823
20075255 28648823 20566863 28004739 24508170 25474469 17510324 18362917 19820115 23537049 25830018 24495485 25315136 12364791 14747013 17210077 26823975 26108605 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 12537569 23723238 25512509 27672113 27166823
EMBL
NWSH01000730
PCG74553.1
PCG74555.1
PCG74552.1
PCG74554.1
RSAL01000003
+ More
RVE54847.1 PCG74551.1 GDQN01006869 JAT84185.1 AGBW02012322 OWR45226.1 GDQN01007419 JAT83635.1 KQ461175 KPJ07848.1 KQ459302 KPJ01766.1 BABH01024477 BABH01024478 BABH01024479 GL446901 EFN87095.1 ADTU01005761 ADTU01005762 ADTU01005763 ADTU01005764 ADTU01005765 ADTU01005766 ADTU01005767 KQ414661 KOC65522.1 KZ288230 PBC31633.1 KQ435855 KOX70637.1 QOIP01000001 RLU26392.1 AAZX01006262 GL439108 EFN67807.1 NNAY01000061 OXU31405.1 KQ434899 KZC10817.1 KQ982691 KYQ52041.1 DS235763 EEB16663.1 GEZM01088826 JAV57876.1 GEZM01088831 JAV57870.1 GFDF01006419 JAV07665.1 KQ981200 KYN45026.1 GFDL01009194 JAV25851.1 KK107260 EZA54070.1 GBBI01003463 JAC15249.1 GFDL01009188 JAV25857.1 GEDC01030206 GEDC01013922 JAS07092.1 JAS23376.1 DS232000 EDS31142.1 CH477944 EAT34884.1 KQ971371 EFA09893.1 GGFK01004942 MBW38263.1 APGK01043371 APGK01043372 APGK01043373 KB741014 ENN75447.1 GBXI01007420 JAD06872.1 GAMC01013315 JAB93240.1 AAAB01008807 EAA45449.4 GDHC01003374 JAQ15255.1 CCAG010014092 CCAG010014093 GECZ01019853 JAS49916.1 GEZM01088836 JAV57860.1 GAMC01013313 JAB93242.1 JRES01000254 KNC32927.1 GEZM01088830 JAV57871.1 CVRI01000059 CRL03427.1 GEDC01028514 JAS08784.1 GECZ01000571 JAS69198.1 CRL03426.1 CH954178 KQS43078.1 KQS43074.1 GBXI01007145 JAD07147.1 AJWK01021990 AJWK01021991 GECZ01025621 JAS44148.1 CH379067 EAL31308.3 CH933816 KRG07618.1 CH940647 KRF84042.1 AE014296 AGB93955.1 CM000159 KRK00748.1 EDV50171.2 CM002912 KMY96843.1 KQS43082.1 KRG07617.1 KRT07390.1 KRT07392.1 KRF84044.1 KRK00749.1 OUUW01000009 SPP84863.1 EDW92846.2 CH902618 KPU77708.1 CH963847 EDW72802.2 AY075414 BT125948 AAL68237.1 ADX35927.1 AAN11486.1 KPU77717.1 KMY96845.1 KMY96844.1 EDW17128.2 KQS43081.1
RVE54847.1 PCG74551.1 GDQN01006869 JAT84185.1 AGBW02012322 OWR45226.1 GDQN01007419 JAT83635.1 KQ461175 KPJ07848.1 KQ459302 KPJ01766.1 BABH01024477 BABH01024478 BABH01024479 GL446901 EFN87095.1 ADTU01005761 ADTU01005762 ADTU01005763 ADTU01005764 ADTU01005765 ADTU01005766 ADTU01005767 KQ414661 KOC65522.1 KZ288230 PBC31633.1 KQ435855 KOX70637.1 QOIP01000001 RLU26392.1 AAZX01006262 GL439108 EFN67807.1 NNAY01000061 OXU31405.1 KQ434899 KZC10817.1 KQ982691 KYQ52041.1 DS235763 EEB16663.1 GEZM01088826 JAV57876.1 GEZM01088831 JAV57870.1 GFDF01006419 JAV07665.1 KQ981200 KYN45026.1 GFDL01009194 JAV25851.1 KK107260 EZA54070.1 GBBI01003463 JAC15249.1 GFDL01009188 JAV25857.1 GEDC01030206 GEDC01013922 JAS07092.1 JAS23376.1 DS232000 EDS31142.1 CH477944 EAT34884.1 KQ971371 EFA09893.1 GGFK01004942 MBW38263.1 APGK01043371 APGK01043372 APGK01043373 KB741014 ENN75447.1 GBXI01007420 JAD06872.1 GAMC01013315 JAB93240.1 AAAB01008807 EAA45449.4 GDHC01003374 JAQ15255.1 CCAG010014092 CCAG010014093 GECZ01019853 JAS49916.1 GEZM01088836 JAV57860.1 GAMC01013313 JAB93242.1 JRES01000254 KNC32927.1 GEZM01088830 JAV57871.1 CVRI01000059 CRL03427.1 GEDC01028514 JAS08784.1 GECZ01000571 JAS69198.1 CRL03426.1 CH954178 KQS43078.1 KQS43074.1 GBXI01007145 JAD07147.1 AJWK01021990 AJWK01021991 GECZ01025621 JAS44148.1 CH379067 EAL31308.3 CH933816 KRG07618.1 CH940647 KRF84042.1 AE014296 AGB93955.1 CM000159 KRK00748.1 EDV50171.2 CM002912 KMY96843.1 KQS43082.1 KRG07617.1 KRT07390.1 KRT07392.1 KRF84044.1 KRK00749.1 OUUW01000009 SPP84863.1 EDW92846.2 CH902618 KPU77708.1 CH963847 EDW72802.2 AY075414 BT125948 AAL68237.1 ADX35927.1 AAN11486.1 KPU77717.1 KMY96845.1 KMY96844.1 EDW17128.2 KQS43081.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
UP000005204
+ More
UP000008237 UP000005205 UP000053825 UP000242457 UP000053105 UP000005203 UP000279307 UP000002358 UP000000311 UP000215335 UP000076502 UP000075809 UP000009046 UP000078541 UP000053097 UP000002320 UP000008820 UP000007266 UP000069272 UP000075920 UP000019118 UP000075903 UP000095301 UP000007062 UP000092444 UP000037069 UP000092445 UP000078200 UP000183832 UP000095300 UP000008711 UP000092461 UP000001819 UP000009192 UP000008792 UP000000803 UP000002282 UP000192221 UP000268350 UP000007801 UP000007798
UP000008237 UP000005205 UP000053825 UP000242457 UP000053105 UP000005203 UP000279307 UP000002358 UP000000311 UP000215335 UP000076502 UP000075809 UP000009046 UP000078541 UP000053097 UP000002320 UP000008820 UP000007266 UP000069272 UP000075920 UP000019118 UP000075903 UP000095301 UP000007062 UP000092444 UP000037069 UP000092445 UP000078200 UP000183832 UP000095300 UP000008711 UP000092461 UP000001819 UP000009192 UP000008792 UP000000803 UP000002282 UP000192221 UP000268350 UP000007801 UP000007798
Pfam
Interpro
IPR036388
WH-like_DNA-bd_sf
+ More
IPR027244 IML1
IPR036390 WH_DNA-bd_sf
IPR000591 DEP_dom
IPR041565 Parkin_Znf-RING
IPR000626 Ubiquitin_dom
IPR036291 NAD(P)-bd_dom_sf
IPR020904 Sc_DH/Rdtase_CS
IPR029071 Ubiquitin-like_domsf
IPR041170 Znf-RING_14
IPR003977 Parkin
IPR002867 IBR_dom
IPR000182 GNAT_dom
IPR016181 Acyl_CoA_acyltransferase
IPR000905 Gcp-like_dom
IPR027244 IML1
IPR036390 WH_DNA-bd_sf
IPR000591 DEP_dom
IPR041565 Parkin_Znf-RING
IPR000626 Ubiquitin_dom
IPR036291 NAD(P)-bd_dom_sf
IPR020904 Sc_DH/Rdtase_CS
IPR029071 Ubiquitin-like_domsf
IPR041170 Znf-RING_14
IPR003977 Parkin
IPR002867 IBR_dom
IPR000182 GNAT_dom
IPR016181 Acyl_CoA_acyltransferase
IPR000905 Gcp-like_dom
Gene 3D
ProteinModelPortal
A0A2A4JSW6
A0A2A4JR85
A0A2A4JT36
A0A2A4JSG8
A0A3S2P8B4
A0A2A4JRD6
+ More
A0A1E1WB64 A0A212EUR6 A0A1E1W9K7 A0A194QQR6 A0A194QE74 H9JBL4 E2BB04 A0A158P0Q8 A0A0L7R3R3 A0A2A3EIL1 A0A0M8ZTI0 A0A088A8W3 A0A3L8E1I9 K7IT54 E2AFB4 A0A232FL67 A0A154PG92 A0A151WWI1 E0VTF7 A0A1Y1KEN3 A0A1Y1KCF9 A0A1L8DMQ9 A0A195FX74 A0A1Q3FE62 A0A026WDB7 A0A023F2J6 A0A1S4FXU9 A0A1Q3FE70 A0A1B6DCJ2 B0WMQ7 Q16KP8 D6X293 A0A182FDQ0 A0A182WL50 A0A2M4ABU9 N6U9Z9 A0A182UW39 A0A0A1X6Z1 W8AWB3 A0A1I8MH41 Q7PF61 A0A146M7J6 A0A1B0FMG6 A0A1B6FI79 A0A1I8MH47 A0A1Y1KCE5 W8B3W3 A0A0L0CL22 A0A1I8MH44 A0A1A9ZU36 A0A1A9V409 A0A1Y1KBU7 A0A1J1IT55 A0A1B6C5V5 A0A1I8MH54 A0A1I8PYI3 A0A1B6H3H8 A0A1I8MH43 A0A1I8MH46 A0A1J1IUP4 A0A0Q5UGY3 A0A0Q5UH86 A0A1I8MH49 A0A0A1X7M9 A0A1B0GJN7 A0A1B6F1V2 Q29FE7 A0A0Q9XR66 A0A0Q9WHT5 M9PDV3 A0A0R1DUA6 A0A1W4W610 B3NBB8 A0A0J9RLN3 A0A0Q5UGT4 A0A0Q9XT29 A0A0R3P2I6 A0A0R3P7M6 A0A0Q9WGV2 A0A0R1DUJ8 A0A3B0JUX3 B4PD98 A0A0N8P0P6 B4MKU6 Q9W0E3 Q8IRG8 A0A0P9AIP1 A0A1W4W6K3 A0A0J9UC37 A0A1W4VTZ7 A0A0J9RN56 B4L8V3 A0A0Q5U2Y5
A0A1E1WB64 A0A212EUR6 A0A1E1W9K7 A0A194QQR6 A0A194QE74 H9JBL4 E2BB04 A0A158P0Q8 A0A0L7R3R3 A0A2A3EIL1 A0A0M8ZTI0 A0A088A8W3 A0A3L8E1I9 K7IT54 E2AFB4 A0A232FL67 A0A154PG92 A0A151WWI1 E0VTF7 A0A1Y1KEN3 A0A1Y1KCF9 A0A1L8DMQ9 A0A195FX74 A0A1Q3FE62 A0A026WDB7 A0A023F2J6 A0A1S4FXU9 A0A1Q3FE70 A0A1B6DCJ2 B0WMQ7 Q16KP8 D6X293 A0A182FDQ0 A0A182WL50 A0A2M4ABU9 N6U9Z9 A0A182UW39 A0A0A1X6Z1 W8AWB3 A0A1I8MH41 Q7PF61 A0A146M7J6 A0A1B0FMG6 A0A1B6FI79 A0A1I8MH47 A0A1Y1KCE5 W8B3W3 A0A0L0CL22 A0A1I8MH44 A0A1A9ZU36 A0A1A9V409 A0A1Y1KBU7 A0A1J1IT55 A0A1B6C5V5 A0A1I8MH54 A0A1I8PYI3 A0A1B6H3H8 A0A1I8MH43 A0A1I8MH46 A0A1J1IUP4 A0A0Q5UGY3 A0A0Q5UH86 A0A1I8MH49 A0A0A1X7M9 A0A1B0GJN7 A0A1B6F1V2 Q29FE7 A0A0Q9XR66 A0A0Q9WHT5 M9PDV3 A0A0R1DUA6 A0A1W4W610 B3NBB8 A0A0J9RLN3 A0A0Q5UGT4 A0A0Q9XT29 A0A0R3P2I6 A0A0R3P7M6 A0A0Q9WGV2 A0A0R1DUJ8 A0A3B0JUX3 B4PD98 A0A0N8P0P6 B4MKU6 Q9W0E3 Q8IRG8 A0A0P9AIP1 A0A1W4W6K3 A0A0J9UC37 A0A1W4VTZ7 A0A0J9RN56 B4L8V3 A0A0Q5U2Y5
PDB
6CET
E-value=0,
Score=2021
Ontologies
GO
PANTHER
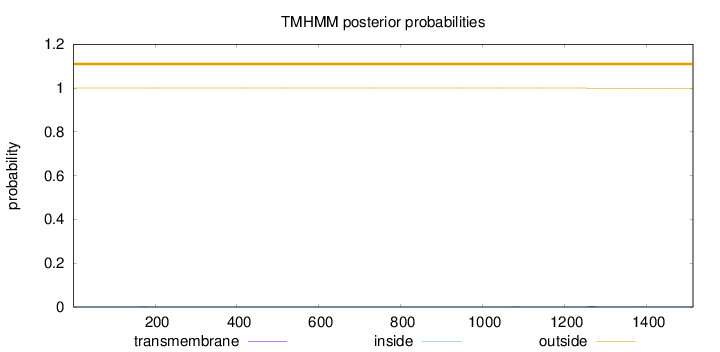
Topology
Length:
1513
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0675400000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00031
outside
1 - 1513
Population Genetic Test Statistics
Pi
201.534252
Theta
157.623861
Tajima's D
0.942118
CLR
0.132598
CSRT
0.63896805159742
Interpretation
Uncertain
