Pre Gene Modal
BGIBMGA006946
Annotation
caspase-1_[Bombyx_mori]
Full name
Caspase-1
+ More
Caspase
Caspase
Alternative Name
drICE
Location in the cell
Extracellular Reliability : 1.785 Nuclear Reliability : 1.308
Sequence
CDS
ATGGCTGATGAAGAAAAGAAAAACAATGGCTCTGGAACAGAACAAAGGAAAAACGGCAACGAAGACGAAGGCGACGCATGGGGCAGTCATGGATCTTCACAAGGGCGACGTTATGCGAAAATGCCTGTCGAAAGATATGCTCATTACTATAACATGAACCACAATAATCGTGGTATGGCGATCATTTTCAATCATGAACATTTTGAAATTCATAATCTAAAATCTCGCACGGGCACCAATGTGGACAGTGACAGCTTATCTAAAGTCCTGAGAGGCTTAGGTTTCAGTGTTACTGTTCTTCATAACCTAAGAGCTGAAGATATCAACAGATATATACAACAGATATCTGAAATGGATCACACAGATAATGATTGTCTGCTGGTTGCTGTGTTGAGTCATGGAGAATTAGGTATGTTGTATGCCAAAGACACTCATTATAAACCAGATAATCTGTGGTATTACTTCACTGCTGATAAATGTCCAACTCTAGCTGGAAAACCAAAGCTGTTCTTTATTCAAGCTTGCCAAGGTGATAAGTTAGATGGTGGAATAACATTATCCAACACTGAAACAGATGGTTCTTCGTCTTCATCCTACAGAATCCCAATTCATGCTGACTTTTTGATTGTGTTCTCAACGGTACCAGGTTATTACTCTTGGAGAAACACAACTCGTGGCTCTTGGTTTATGCAATCTCTTTGTGAAGAATTGAGAAACTATGGTACTCAAAGAGATATTCTTACACTCTTGACATTTGTTTGTCAGAGAGTTGCTTTAGATTTTGAGTCAAATACACCAGATATCACACCGATGCATCAACAAAAACAGGTACCTTGCATAAATAGTATGCTGACACGCCTTCTCTTGTTTGGAAAAAAATAA
Protein
MADEEKKNNGSGTEQRKNGNEDEGDAWGSHGSSQGRRYAKMPVERYAHYYNMNHNNRGMAIIFNHEHFEIHNLKSRTGTNVDSDSLSKVLRGLGFSVTVLHNLRAEDINRYIQQISEMDHTDNDCLLVAVLSHGELGMLYAKDTHYKPDNLWYYFTADKCPTLAGKPKLFFIQACQGDKLDGGITLSNTETDGSSSSSYRIPIHADFLIVFSTVPGYYSWRNTTRGSWFMQSLCEELRNYGTQRDILTLLTFVCQRVALDFESNTPDITPMHQQKQVPCINSMLTRLLLFGKK
Summary
Description
Involved in the activation cascade of caspases responsible for apoptosis execution (By similarity). Inhibited by the baculovirus anti-apoptotic protein p35. Cleaves p35 and nuclear immunophilin FKBP46.
Subunit
Heterotetramer that consists of two anti-parallel arranged heterodimers, each one formed by a 19/18 kDa (p19/18) and a 12 kDa (p12) subunit.
Heterotetramer that consists of two anti-parallel arranged heterodimers, each one formed by a 21 kDa (p21) and a 12 kDa (p12) subunit. Inactive pro-form can homodimerize. Dronc and Drice can form a stable complex (PubMed:10675329). Interacts with Diap2 (via BIR3 domain) to form a stable complex (PubMed:18166655).
Heterotetramer that consists of two anti-parallel arranged heterodimers, each one formed by a 21 kDa (p21) and a 12 kDa (p12) subunit. Inactive pro-form can homodimerize. Dronc and Drice can form a stable complex (PubMed:10675329). Interacts with Diap2 (via BIR3 domain) to form a stable complex (PubMed:18166655).
Similarity
Belongs to the peptidase C14A family.
Keywords
3D-structure
Apoptosis
Direct protein sequencing
Hydrolase
Protease
Thiol protease
Zymogen
Complete proteome
Reference proteome
Feature
chain Caspase-1 subunit p19/18
Uniprot
H9JBQ1
Q8I9V7
F6K5R8
A0A1E1WPY0
B6EEC1
I2EC37
+ More
Q2TVJ7 F6K5S3 A7L9Z3 G1ELA2 A9Q0J3 G0XQE9 C6YXH1 G0XQE8 A0A0L7KY59 K4PXX6 H6V6K4 Q8I955 A0A194QQQ6 A0A2A4JRA2 A0A194Q8Q5 A0A2H1VLH4 P89116 D9IVD4 S4PEK3 G0XQE5 J9PJR5 A0A212EUQ8 G5CV11 G0XQE6 K4Q332 G0XQE4 F8UU16 G0XQE7 A0A0K8TSG1 A0A1W4W9I1 K7J065 A0A232EYP6 A0A1W4W9I8 A0A154PEN4 O01382 A0A1B0DJJ8 B4HZF1 A0A336MXM6 B4R093 A0A0C9RND9 A0A1Q3EXQ9 A0A3L8DWL7 E2AAJ3 B4NBM1 B0W0K2 A0A1W4UCU4 B4KAN6 B4PPJ9 D6WFK4 B4M0P7 A0A139WKB8 A0A1B0G224 A0A1P7XV02 E2B4X5 D3TL84 A0A182PVZ2 A0A2J7PIQ1 A0A1A9ZSW0 A0A182XXX5 A0A1A9VDR7 A0A182RMT7 A0A088A747 A0A1B0B2Z4 F4WFL7 A0A1A9W5Q4 A0A3B0JG01 A0A1L2F0C2 A0A182H4R2 Q297T5 A0A182LYX6 A0A0M4EUT4 B3LWX5 B4G307 A0A151J4A4 A0A1I8M9Z6 A0A084VH01 A0A182Q649 A0A1J1IZC7 E9IB18 A0A182W5E0 A0A182N1J2 T1EA88 A0A182IX37 A0A151HYK2 A0A0J7N6M5 B5AK94 A0A1I8P6R3 A0A151WIC1 B4JRJ9 W5JK07 A0A2M3ZJ43 A0A2M4AW77 A0A1W4UNE7 B3NPN7
Q2TVJ7 F6K5S3 A7L9Z3 G1ELA2 A9Q0J3 G0XQE9 C6YXH1 G0XQE8 A0A0L7KY59 K4PXX6 H6V6K4 Q8I955 A0A194QQQ6 A0A2A4JRA2 A0A194Q8Q5 A0A2H1VLH4 P89116 D9IVD4 S4PEK3 G0XQE5 J9PJR5 A0A212EUQ8 G5CV11 G0XQE6 K4Q332 G0XQE4 F8UU16 G0XQE7 A0A0K8TSG1 A0A1W4W9I1 K7J065 A0A232EYP6 A0A1W4W9I8 A0A154PEN4 O01382 A0A1B0DJJ8 B4HZF1 A0A336MXM6 B4R093 A0A0C9RND9 A0A1Q3EXQ9 A0A3L8DWL7 E2AAJ3 B4NBM1 B0W0K2 A0A1W4UCU4 B4KAN6 B4PPJ9 D6WFK4 B4M0P7 A0A139WKB8 A0A1B0G224 A0A1P7XV02 E2B4X5 D3TL84 A0A182PVZ2 A0A2J7PIQ1 A0A1A9ZSW0 A0A182XXX5 A0A1A9VDR7 A0A182RMT7 A0A088A747 A0A1B0B2Z4 F4WFL7 A0A1A9W5Q4 A0A3B0JG01 A0A1L2F0C2 A0A182H4R2 Q297T5 A0A182LYX6 A0A0M4EUT4 B3LWX5 B4G307 A0A151J4A4 A0A1I8M9Z6 A0A084VH01 A0A182Q649 A0A1J1IZC7 E9IB18 A0A182W5E0 A0A182N1J2 T1EA88 A0A182IX37 A0A151HYK2 A0A0J7N6M5 B5AK94 A0A1I8P6R3 A0A151WIC1 B4JRJ9 W5JK07 A0A2M3ZJ43 A0A2M4AW77 A0A1W4UNE7 B3NPN7
EC Number
3.4.22.-
Pubmed
19121390
12324475
21740565
19027856
17541728
17603110
+ More
26227816 26354079 8999805 23622113 22118469 22229544 26369729 20075255 28648823 9184225 10731132 12537572 12537569 10675329 18166655 17994087 30249741 20798317 17550304 18362917 19820115 20353571 25244985 21719571 26483478 15632085 25315136 24438588 21282665 20920257 23761445
26227816 26354079 8999805 23622113 22118469 22229544 26369729 20075255 28648823 9184225 10731132 12537572 12537569 10675329 18166655 17994087 30249741 20798317 17550304 18362917 19820115 20353571 25244985 21719571 26483478 15632085 25315136 24438588 21282665 20920257 23761445
EMBL
BABH01024476
AF448494
AAN86250.1
HM234675
AEF30493.1
GDQN01002016
+ More
JAT89038.1 FJ266474 ACI43910.1 JQ864206 AFJ97219.1 AY159381 AAO17788.1 HM234680 AEF30498.1 EF688063 HQ645847 ABS18284.1 AEH16632.1 JN113041 AEK12771.1 EF190236 ABO93468.1 HQ328953 AEK20810.1 EF211965 ACU11588.1 HQ328952 AEK20809.1 JTDY01004421 KOB68178.1 AB755808 BAM62940.1 JN794535 AFA43940.1 AF548387 AAO16241.1 KQ461175 KPJ07838.1 NWSH01000730 PCG74557.1 KQ459302 KPJ01774.1 ODYU01003219 SOQ41673.1 U81510 HM204505 ADK24087.1 GAIX01004462 JAA88098.1 HQ328949 AEK20806.1 JF798683 AEQ18745.1 AGBW02012322 OWR45225.1 JN410830 AEP25406.1 HQ328950 AEK20807.1 AB755807 BAM62939.1 HQ328948 AEK20805.1 JF906728 AEH76885.1 HQ328951 AEK20808.1 GDAI01000733 JAI16870.1 NNAY01001601 OXU23463.1 KQ434878 KZC09894.1 Y12261 AE014297 AY058451 AJVK01028708 AJVK01035047 CH480819 EDW53408.1 UFQT01001457 SSX30638.1 CM000364 EDX14899.1 GBYB01009880 JAG79647.1 GFDL01014948 JAV20097.1 QOIP01000003 RLU24837.1 GL438128 EFN69540.1 CH964232 EDW81185.1 DS231818 EDS41304.1 CH933806 EDW16773.1 CM000160 EDW98249.1 KQ971328 EFA01345.2 CH940650 EDW67339.1 KYB28363.1 CCAG010010758 KF955542 KF955543 KZ288416 AIS23518.1 AIS23519.1 PBC26031.1 GL445671 EFN89257.1 EZ422185 ADD18379.1 NEVH01024980 PNF16221.1 JXJN01007738 GL888120 EGI66994.1 OUUW01000005 SPP81055.1 KX463639 ANQ91917.1 JXUM01003922 JXUM01003923 KQ560165 KXJ84059.1 CM000070 EAL28120.3 AXCM01000181 CP012526 ALC46917.1 CH902617 EDV43814.1 CH479179 EDW24202.1 KQ980159 KYN17427.1 ATLV01013118 KE524840 KFB37245.1 AXCN02001449 CVRI01000063 CRL04900.1 GL762102 EFZ22232.1 GAMD01001223 JAB00368.1 KQ976719 KYM76754.1 LBMM01009157 KMQ88345.1 EU854472 ACF71490.1 KQ983089 KYQ47566.1 CH916373 EDV94389.1 ADMH02001211 ETN63643.1 GGFM01007846 MBW28597.1 GGFK01011728 MBW45049.1 CH954179 EDV56828.1
JAT89038.1 FJ266474 ACI43910.1 JQ864206 AFJ97219.1 AY159381 AAO17788.1 HM234680 AEF30498.1 EF688063 HQ645847 ABS18284.1 AEH16632.1 JN113041 AEK12771.1 EF190236 ABO93468.1 HQ328953 AEK20810.1 EF211965 ACU11588.1 HQ328952 AEK20809.1 JTDY01004421 KOB68178.1 AB755808 BAM62940.1 JN794535 AFA43940.1 AF548387 AAO16241.1 KQ461175 KPJ07838.1 NWSH01000730 PCG74557.1 KQ459302 KPJ01774.1 ODYU01003219 SOQ41673.1 U81510 HM204505 ADK24087.1 GAIX01004462 JAA88098.1 HQ328949 AEK20806.1 JF798683 AEQ18745.1 AGBW02012322 OWR45225.1 JN410830 AEP25406.1 HQ328950 AEK20807.1 AB755807 BAM62939.1 HQ328948 AEK20805.1 JF906728 AEH76885.1 HQ328951 AEK20808.1 GDAI01000733 JAI16870.1 NNAY01001601 OXU23463.1 KQ434878 KZC09894.1 Y12261 AE014297 AY058451 AJVK01028708 AJVK01035047 CH480819 EDW53408.1 UFQT01001457 SSX30638.1 CM000364 EDX14899.1 GBYB01009880 JAG79647.1 GFDL01014948 JAV20097.1 QOIP01000003 RLU24837.1 GL438128 EFN69540.1 CH964232 EDW81185.1 DS231818 EDS41304.1 CH933806 EDW16773.1 CM000160 EDW98249.1 KQ971328 EFA01345.2 CH940650 EDW67339.1 KYB28363.1 CCAG010010758 KF955542 KF955543 KZ288416 AIS23518.1 AIS23519.1 PBC26031.1 GL445671 EFN89257.1 EZ422185 ADD18379.1 NEVH01024980 PNF16221.1 JXJN01007738 GL888120 EGI66994.1 OUUW01000005 SPP81055.1 KX463639 ANQ91917.1 JXUM01003922 JXUM01003923 KQ560165 KXJ84059.1 CM000070 EAL28120.3 AXCM01000181 CP012526 ALC46917.1 CH902617 EDV43814.1 CH479179 EDW24202.1 KQ980159 KYN17427.1 ATLV01013118 KE524840 KFB37245.1 AXCN02001449 CVRI01000063 CRL04900.1 GL762102 EFZ22232.1 GAMD01001223 JAB00368.1 KQ976719 KYM76754.1 LBMM01009157 KMQ88345.1 EU854472 ACF71490.1 KQ983089 KYQ47566.1 CH916373 EDV94389.1 ADMH02001211 ETN63643.1 GGFM01007846 MBW28597.1 GGFK01011728 MBW45049.1 CH954179 EDV56828.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000218220
UP000053268
UP000007151
+ More
UP000192223 UP000002358 UP000215335 UP000076502 UP000000803 UP000092462 UP000001292 UP000000304 UP000279307 UP000000311 UP000007798 UP000002320 UP000192221 UP000009192 UP000002282 UP000007266 UP000008792 UP000092444 UP000242457 UP000008237 UP000075885 UP000235965 UP000092445 UP000076408 UP000078200 UP000075900 UP000005203 UP000092460 UP000007755 UP000091820 UP000268350 UP000069940 UP000249989 UP000001819 UP000075883 UP000092553 UP000007801 UP000008744 UP000078492 UP000095301 UP000030765 UP000075886 UP000183832 UP000075920 UP000075884 UP000075880 UP000078540 UP000036403 UP000095300 UP000075809 UP000001070 UP000000673 UP000008711
UP000192223 UP000002358 UP000215335 UP000076502 UP000000803 UP000092462 UP000001292 UP000000304 UP000279307 UP000000311 UP000007798 UP000002320 UP000192221 UP000009192 UP000002282 UP000007266 UP000008792 UP000092444 UP000242457 UP000008237 UP000075885 UP000235965 UP000092445 UP000076408 UP000078200 UP000075900 UP000005203 UP000092460 UP000007755 UP000091820 UP000268350 UP000069940 UP000249989 UP000001819 UP000075883 UP000092553 UP000007801 UP000008744 UP000078492 UP000095301 UP000030765 UP000075886 UP000183832 UP000075920 UP000075884 UP000075880 UP000078540 UP000036403 UP000095300 UP000075809 UP000001070 UP000000673 UP000008711
Interpro
SUPFAM
SSF52129
SSF52129
CDD
ProteinModelPortal
H9JBQ1
Q8I9V7
F6K5R8
A0A1E1WPY0
B6EEC1
I2EC37
+ More
Q2TVJ7 F6K5S3 A7L9Z3 G1ELA2 A9Q0J3 G0XQE9 C6YXH1 G0XQE8 A0A0L7KY59 K4PXX6 H6V6K4 Q8I955 A0A194QQQ6 A0A2A4JRA2 A0A194Q8Q5 A0A2H1VLH4 P89116 D9IVD4 S4PEK3 G0XQE5 J9PJR5 A0A212EUQ8 G5CV11 G0XQE6 K4Q332 G0XQE4 F8UU16 G0XQE7 A0A0K8TSG1 A0A1W4W9I1 K7J065 A0A232EYP6 A0A1W4W9I8 A0A154PEN4 O01382 A0A1B0DJJ8 B4HZF1 A0A336MXM6 B4R093 A0A0C9RND9 A0A1Q3EXQ9 A0A3L8DWL7 E2AAJ3 B4NBM1 B0W0K2 A0A1W4UCU4 B4KAN6 B4PPJ9 D6WFK4 B4M0P7 A0A139WKB8 A0A1B0G224 A0A1P7XV02 E2B4X5 D3TL84 A0A182PVZ2 A0A2J7PIQ1 A0A1A9ZSW0 A0A182XXX5 A0A1A9VDR7 A0A182RMT7 A0A088A747 A0A1B0B2Z4 F4WFL7 A0A1A9W5Q4 A0A3B0JG01 A0A1L2F0C2 A0A182H4R2 Q297T5 A0A182LYX6 A0A0M4EUT4 B3LWX5 B4G307 A0A151J4A4 A0A1I8M9Z6 A0A084VH01 A0A182Q649 A0A1J1IZC7 E9IB18 A0A182W5E0 A0A182N1J2 T1EA88 A0A182IX37 A0A151HYK2 A0A0J7N6M5 B5AK94 A0A1I8P6R3 A0A151WIC1 B4JRJ9 W5JK07 A0A2M3ZJ43 A0A2M4AW77 A0A1W4UNE7 B3NPN7
Q2TVJ7 F6K5S3 A7L9Z3 G1ELA2 A9Q0J3 G0XQE9 C6YXH1 G0XQE8 A0A0L7KY59 K4PXX6 H6V6K4 Q8I955 A0A194QQQ6 A0A2A4JRA2 A0A194Q8Q5 A0A2H1VLH4 P89116 D9IVD4 S4PEK3 G0XQE5 J9PJR5 A0A212EUQ8 G5CV11 G0XQE6 K4Q332 G0XQE4 F8UU16 G0XQE7 A0A0K8TSG1 A0A1W4W9I1 K7J065 A0A232EYP6 A0A1W4W9I8 A0A154PEN4 O01382 A0A1B0DJJ8 B4HZF1 A0A336MXM6 B4R093 A0A0C9RND9 A0A1Q3EXQ9 A0A3L8DWL7 E2AAJ3 B4NBM1 B0W0K2 A0A1W4UCU4 B4KAN6 B4PPJ9 D6WFK4 B4M0P7 A0A139WKB8 A0A1B0G224 A0A1P7XV02 E2B4X5 D3TL84 A0A182PVZ2 A0A2J7PIQ1 A0A1A9ZSW0 A0A182XXX5 A0A1A9VDR7 A0A182RMT7 A0A088A747 A0A1B0B2Z4 F4WFL7 A0A1A9W5Q4 A0A3B0JG01 A0A1L2F0C2 A0A182H4R2 Q297T5 A0A182LYX6 A0A0M4EUT4 B3LWX5 B4G307 A0A151J4A4 A0A1I8M9Z6 A0A084VH01 A0A182Q649 A0A1J1IZC7 E9IB18 A0A182W5E0 A0A182N1J2 T1EA88 A0A182IX37 A0A151HYK2 A0A0J7N6M5 B5AK94 A0A1I8P6R3 A0A151WIC1 B4JRJ9 W5JK07 A0A2M3ZJ43 A0A2M4AW77 A0A1W4UNE7 B3NPN7
PDB
2NN3
E-value=1.47847e-136,
Score=1244
Ontologies
PATHWAY
GO
PANTHER
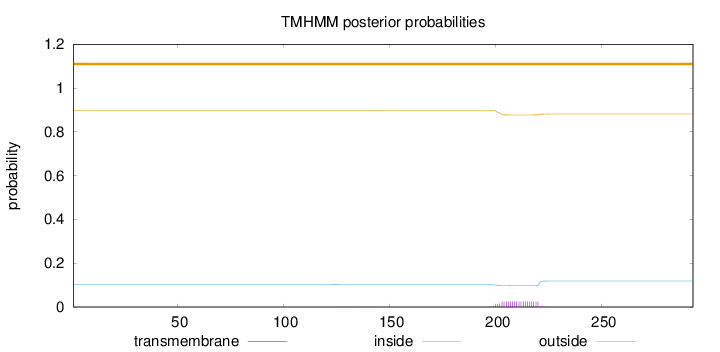
Topology
Length:
293
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.505399999999999
Exp number, first 60 AAs:
0.00044
Total prob of N-in:
0.10255
outside
1 - 293
Population Genetic Test Statistics
Pi
235.723211
Theta
154.665156
Tajima's D
1.887408
CLR
1.517568
CSRT
0.861906904654767
Interpretation
Possibly Positive selection
