Gene
KWMTBOMO05809
Pre Gene Modal
BGIBMGA006944
Annotation
PREDICTED:_protein_Asterix_[Amyelois_transitella]
Full name
Protein Asterix
Alternative Name
Protein WDR83OS homolog
Location in the cell
Nuclear Reliability : 1.537 PlasmaMembrane Reliability : 1.775
Sequence
CDS
ATGCAGTTGTCAGCAGATCCACGTCGCGCTGACAGAGAACGTCGCTATAAACCACCTCCACCCACTTCTGCTCCAGCAGAAGACCTAACCACAGATTATATGAATATTTTAGGGATGGTATTTTCAATGTGTGGCTTAATGATGAGATTGAAATGGTGCGCGTGGACAGCTGTATTTTGTTCCAGCATCAGTTTTGCAAACTCAAGAGTTTCCGATGACACTAAACAGATTGTGAGTTCCTTCATGTTGTCCATTTCAGCAGTAGTAATGTCATATCTTCAAAACCCTGCACCAATGACACCACCTTGGGCAGCTCTTACTACATGA
Protein
MQLSADPRRADRERRYKPPPPTSAPAEDLTTDYMNILGMVFSMCGLMMRLKWCAWTAVFCSSISFANSRVSDDTKQIVSSFMLSISAVVMSYLQNPAPMTPPWAALTT
Summary
Similarity
Belongs to the Asterix family.
Keywords
Membrane
Transmembrane
Transmembrane helix
Complete proteome
Reference proteome
Feature
chain Protein Asterix
Uniprot
A0A2A4JSV6
A0A2H1WI86
S4PLX4
F8SL46
A0A1E1WQX4
I4DJB2
+ More
A0A194QW96 Q9U516 A0A194Q9V2 A0A0L7K4K9 A0A0L7KYS6 A0A2R7X0P6 A0A0P4VRS5 R4G3E7 A0A232FCH3 K7J7J1 A0A224XQ33 A0A0V0G7J5 A0A069DND8 A0A1A9X0I7 A0A0L7R049 A0A1A9XIM7 A0A1B0AQD6 A0A1A9ZMI2 A0A1B0FAP4 A0A154PKA1 E2C9C5 A0A2A3ELJ2 A0A088AKA6 A0A067QUU5 A0A1A9VN16 A0A1B6I4F9 A0A1B6GH45 A0A0A9WLQ0 A0A0N0U4Y9 A0A1S3DDQ4 A0A0J7K5K3 A0A026WWI1 E2AMV8 A0A2P8Z694 A0A0L0BL52 T1D5H0 A0A0B6Z186 A0A034WJC3 A0A0C9SA70 A0A1E1WXD1 L7M2G9 W8BTT0 A0A0P6IUS3 A0A023FG34 A0A023FTF8 A0A1S3GLL4 A0A1I8Q1W6 A0A1S3ABQ7 A0A1L8EEF7 A0A1I8N996 A0A0K8UQ58 T1E8T5 W5JV84 A0A2M3ZBV2 A0A2M4AP52 A0A182FCJ7 G3MLR4 A0A023GHM2 A0A131XCJ5 A0A224Z3I7 A0A131YQF7 U3FPK9 A0A023EEY4 A0A182JJE6 E9IX33 H9H1M0 F8RT80 A0A1Q3FXM5 B0XED5 A0A2Y9EA22 A0A1A6H5Z3 I3M8N9 A0A250Y928 H0WCU5 A0A091CL71 B2GV64 G3HCV5 G5BYY0 A0A2K6EDD2 A0A2K6K9E0 A0A384DSD7 G1PKY9 G3RC27 F1PKC8 G1RQA1 A0A2K5Y946 A0A0D9R2R2 A0A2U3V3P2 A0A3Q7NKN9 A0A340Y658 A0A2R8ZEE2 A0A0A0MVA8 A0A2K6Q285 A0A2Y9IFB2
A0A194QW96 Q9U516 A0A194Q9V2 A0A0L7K4K9 A0A0L7KYS6 A0A2R7X0P6 A0A0P4VRS5 R4G3E7 A0A232FCH3 K7J7J1 A0A224XQ33 A0A0V0G7J5 A0A069DND8 A0A1A9X0I7 A0A0L7R049 A0A1A9XIM7 A0A1B0AQD6 A0A1A9ZMI2 A0A1B0FAP4 A0A154PKA1 E2C9C5 A0A2A3ELJ2 A0A088AKA6 A0A067QUU5 A0A1A9VN16 A0A1B6I4F9 A0A1B6GH45 A0A0A9WLQ0 A0A0N0U4Y9 A0A1S3DDQ4 A0A0J7K5K3 A0A026WWI1 E2AMV8 A0A2P8Z694 A0A0L0BL52 T1D5H0 A0A0B6Z186 A0A034WJC3 A0A0C9SA70 A0A1E1WXD1 L7M2G9 W8BTT0 A0A0P6IUS3 A0A023FG34 A0A023FTF8 A0A1S3GLL4 A0A1I8Q1W6 A0A1S3ABQ7 A0A1L8EEF7 A0A1I8N996 A0A0K8UQ58 T1E8T5 W5JV84 A0A2M3ZBV2 A0A2M4AP52 A0A182FCJ7 G3MLR4 A0A023GHM2 A0A131XCJ5 A0A224Z3I7 A0A131YQF7 U3FPK9 A0A023EEY4 A0A182JJE6 E9IX33 H9H1M0 F8RT80 A0A1Q3FXM5 B0XED5 A0A2Y9EA22 A0A1A6H5Z3 I3M8N9 A0A250Y928 H0WCU5 A0A091CL71 B2GV64 G3HCV5 G5BYY0 A0A2K6EDD2 A0A2K6K9E0 A0A384DSD7 G1PKY9 G3RC27 F1PKC8 G1RQA1 A0A2K5Y946 A0A0D9R2R2 A0A2U3V3P2 A0A3Q7NKN9 A0A340Y658 A0A2R8ZEE2 A0A0A0MVA8 A0A2K6Q285 A0A2Y9IFB2
Pubmed
23622113
21740565
22651552
26354079
10620045
26227816
+ More
27129103 28648823 20075255 26334808 20798317 24845553 25401762 24508170 30249741 29403074 26108605 24330624 25348373 26131772 28503490 25576852 24495485 26999592 25315136 20920257 23761445 22216098 28049606 28797301 26830274 25243066 24945155 26483478 21282665 20838655 21559472 28087693 21993624 15489334 15057822 15632090 21804562 23929341 29704459 21993625 22398555 16341006 22722832 25362486
27129103 28648823 20075255 26334808 20798317 24845553 25401762 24508170 30249741 29403074 26108605 24330624 25348373 26131772 28503490 25576852 24495485 26999592 25315136 20920257 23761445 22216098 28049606 28797301 26830274 25243066 24945155 26483478 21282665 20838655 21559472 28087693 21993624 15489334 15057822 15632090 21804562 23929341 29704459 21993625 22398555 16341006 22722832 25362486
EMBL
NWSH01000730
PCG74543.1
ODYU01008746
SOQ52612.1
GAIX01003815
JAA88745.1
+ More
HQ645847 AEH16629.1 GDQN01001823 JAT89231.1 AK401380 BAM18002.1 KQ461175 KPJ07841.1 AF117572 KQ459302 KPJ01770.1 JTDY01010886 KOB57913.1 JTDY01004421 KOB68179.1 KK856130 PTY24970.1 GDKW01002000 JAI54595.1 ACPB03009795 GAHY01001746 JAA75764.1 NNAY01000470 OXU28148.1 GFTR01001821 JAW14605.1 GECL01002797 JAP03327.1 GBGD01003559 JAC85330.1 KQ414672 KOC64225.1 JXJN01001810 CCAG010020395 KQ434943 KZC12253.1 GL453806 EFN75470.1 KZ288217 PBC32364.1 KK852908 KDR13958.1 GECU01025912 JAS81794.1 GECZ01030464 GECZ01008001 JAS39305.1 JAS61768.1 GBHO01034930 JAG08674.1 KQ435808 KOX72978.1 LBMM01013500 KMQ85612.1 KK107105 QOIP01000011 EZA59484.1 RLU17059.1 GL440936 EFN65232.1 PYGN01000175 PSN52025.1 JRES01001708 KNC20678.1 GALA01000513 JAA94339.1 HACG01015322 CEK62187.1 GAKP01004752 JAC54200.1 GBZX01002783 JAG89957.1 GFAC01007544 JAT91644.1 GACK01006513 JAA58521.1 GAMC01001795 JAC04761.1 GDUN01000403 JAN95516.1 GBBK01004409 GBBK01004408 GBBK01004407 JAC20075.1 GBBL01002373 JAC24947.1 GFDG01001784 JAV17015.1 GDHF01023507 JAI28807.1 GAMD01002478 JAA99112.1 ADMH02000350 GGFL01004484 ETN66870.1 MBW68662.1 GGFM01005220 MBW25971.1 GGFK01009244 MBW42565.1 JO842815 AEO34432.1 GBBM01003154 GBBM01003153 JAC32265.1 GEFH01004761 JAP63820.1 GFPF01009648 MAA20794.1 GEDV01007785 JAP80772.1 GAMP01008775 JAB43980.1 JXUM01068552 GAPW01006321 GAPW01006318 GAPW01006317 KQ562505 JAC07280.1 KXJ75773.1 GL766616 EFZ14869.1 HQ184923 GFDL01002737 JAV32308.1 DS232821 EDS25909.1 LZPO01044522 OBS74018.1 AGTP01088432 GFFW01004705 JAV40083.1 AAKN02038626 KN125026 KFO19494.1 AABR07072634 BC166543 CH473972 AAI66543.1 EDL92153.1 JH000286 KE672766 RAZU01000124 EGW00591.1 ERE78952.1 RLQ71329.1 JH172486 GEBF01001206 EHB14491.1 JAO02427.1 AAPE02050984 CABD030111801 AAEX03012407 ADFV01089262 AQIB01141045 AJFE02048596 AHZZ02013596
HQ645847 AEH16629.1 GDQN01001823 JAT89231.1 AK401380 BAM18002.1 KQ461175 KPJ07841.1 AF117572 KQ459302 KPJ01770.1 JTDY01010886 KOB57913.1 JTDY01004421 KOB68179.1 KK856130 PTY24970.1 GDKW01002000 JAI54595.1 ACPB03009795 GAHY01001746 JAA75764.1 NNAY01000470 OXU28148.1 GFTR01001821 JAW14605.1 GECL01002797 JAP03327.1 GBGD01003559 JAC85330.1 KQ414672 KOC64225.1 JXJN01001810 CCAG010020395 KQ434943 KZC12253.1 GL453806 EFN75470.1 KZ288217 PBC32364.1 KK852908 KDR13958.1 GECU01025912 JAS81794.1 GECZ01030464 GECZ01008001 JAS39305.1 JAS61768.1 GBHO01034930 JAG08674.1 KQ435808 KOX72978.1 LBMM01013500 KMQ85612.1 KK107105 QOIP01000011 EZA59484.1 RLU17059.1 GL440936 EFN65232.1 PYGN01000175 PSN52025.1 JRES01001708 KNC20678.1 GALA01000513 JAA94339.1 HACG01015322 CEK62187.1 GAKP01004752 JAC54200.1 GBZX01002783 JAG89957.1 GFAC01007544 JAT91644.1 GACK01006513 JAA58521.1 GAMC01001795 JAC04761.1 GDUN01000403 JAN95516.1 GBBK01004409 GBBK01004408 GBBK01004407 JAC20075.1 GBBL01002373 JAC24947.1 GFDG01001784 JAV17015.1 GDHF01023507 JAI28807.1 GAMD01002478 JAA99112.1 ADMH02000350 GGFL01004484 ETN66870.1 MBW68662.1 GGFM01005220 MBW25971.1 GGFK01009244 MBW42565.1 JO842815 AEO34432.1 GBBM01003154 GBBM01003153 JAC32265.1 GEFH01004761 JAP63820.1 GFPF01009648 MAA20794.1 GEDV01007785 JAP80772.1 GAMP01008775 JAB43980.1 JXUM01068552 GAPW01006321 GAPW01006318 GAPW01006317 KQ562505 JAC07280.1 KXJ75773.1 GL766616 EFZ14869.1 HQ184923 GFDL01002737 JAV32308.1 DS232821 EDS25909.1 LZPO01044522 OBS74018.1 AGTP01088432 GFFW01004705 JAV40083.1 AAKN02038626 KN125026 KFO19494.1 AABR07072634 BC166543 CH473972 AAI66543.1 EDL92153.1 JH000286 KE672766 RAZU01000124 EGW00591.1 ERE78952.1 RLQ71329.1 JH172486 GEBF01001206 EHB14491.1 JAO02427.1 AAPE02050984 CABD030111801 AAEX03012407 ADFV01089262 AQIB01141045 AJFE02048596 AHZZ02013596
Proteomes
UP000218220
UP000053240
UP000053268
UP000037510
UP000015103
UP000215335
+ More
UP000002358 UP000091820 UP000053825 UP000092443 UP000092460 UP000092445 UP000092444 UP000076502 UP000008237 UP000242457 UP000005203 UP000027135 UP000078200 UP000053105 UP000079169 UP000036403 UP000053097 UP000279307 UP000000311 UP000245037 UP000037069 UP000081671 UP000095300 UP000079721 UP000095301 UP000000673 UP000069272 UP000069940 UP000249989 UP000075880 UP000001645 UP000000539 UP000002320 UP000248480 UP000092124 UP000005215 UP000005447 UP000028990 UP000002494 UP000001075 UP000030759 UP000273346 UP000006813 UP000233120 UP000233180 UP000261680 UP000001074 UP000001519 UP000002254 UP000001073 UP000233140 UP000029965 UP000245320 UP000286641 UP000265300 UP000240080 UP000028761 UP000233200 UP000248481
UP000002358 UP000091820 UP000053825 UP000092443 UP000092460 UP000092445 UP000092444 UP000076502 UP000008237 UP000242457 UP000005203 UP000027135 UP000078200 UP000053105 UP000079169 UP000036403 UP000053097 UP000279307 UP000000311 UP000245037 UP000037069 UP000081671 UP000095300 UP000079721 UP000095301 UP000000673 UP000069272 UP000069940 UP000249989 UP000075880 UP000001645 UP000000539 UP000002320 UP000248480 UP000092124 UP000005215 UP000005447 UP000028990 UP000002494 UP000001075 UP000030759 UP000273346 UP000006813 UP000233120 UP000233180 UP000261680 UP000001074 UP000001519 UP000002254 UP000001073 UP000233140 UP000029965 UP000245320 UP000286641 UP000265300 UP000240080 UP000028761 UP000233200 UP000248481
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JSV6
A0A2H1WI86
S4PLX4
F8SL46
A0A1E1WQX4
I4DJB2
+ More
A0A194QW96 Q9U516 A0A194Q9V2 A0A0L7K4K9 A0A0L7KYS6 A0A2R7X0P6 A0A0P4VRS5 R4G3E7 A0A232FCH3 K7J7J1 A0A224XQ33 A0A0V0G7J5 A0A069DND8 A0A1A9X0I7 A0A0L7R049 A0A1A9XIM7 A0A1B0AQD6 A0A1A9ZMI2 A0A1B0FAP4 A0A154PKA1 E2C9C5 A0A2A3ELJ2 A0A088AKA6 A0A067QUU5 A0A1A9VN16 A0A1B6I4F9 A0A1B6GH45 A0A0A9WLQ0 A0A0N0U4Y9 A0A1S3DDQ4 A0A0J7K5K3 A0A026WWI1 E2AMV8 A0A2P8Z694 A0A0L0BL52 T1D5H0 A0A0B6Z186 A0A034WJC3 A0A0C9SA70 A0A1E1WXD1 L7M2G9 W8BTT0 A0A0P6IUS3 A0A023FG34 A0A023FTF8 A0A1S3GLL4 A0A1I8Q1W6 A0A1S3ABQ7 A0A1L8EEF7 A0A1I8N996 A0A0K8UQ58 T1E8T5 W5JV84 A0A2M3ZBV2 A0A2M4AP52 A0A182FCJ7 G3MLR4 A0A023GHM2 A0A131XCJ5 A0A224Z3I7 A0A131YQF7 U3FPK9 A0A023EEY4 A0A182JJE6 E9IX33 H9H1M0 F8RT80 A0A1Q3FXM5 B0XED5 A0A2Y9EA22 A0A1A6H5Z3 I3M8N9 A0A250Y928 H0WCU5 A0A091CL71 B2GV64 G3HCV5 G5BYY0 A0A2K6EDD2 A0A2K6K9E0 A0A384DSD7 G1PKY9 G3RC27 F1PKC8 G1RQA1 A0A2K5Y946 A0A0D9R2R2 A0A2U3V3P2 A0A3Q7NKN9 A0A340Y658 A0A2R8ZEE2 A0A0A0MVA8 A0A2K6Q285 A0A2Y9IFB2
A0A194QW96 Q9U516 A0A194Q9V2 A0A0L7K4K9 A0A0L7KYS6 A0A2R7X0P6 A0A0P4VRS5 R4G3E7 A0A232FCH3 K7J7J1 A0A224XQ33 A0A0V0G7J5 A0A069DND8 A0A1A9X0I7 A0A0L7R049 A0A1A9XIM7 A0A1B0AQD6 A0A1A9ZMI2 A0A1B0FAP4 A0A154PKA1 E2C9C5 A0A2A3ELJ2 A0A088AKA6 A0A067QUU5 A0A1A9VN16 A0A1B6I4F9 A0A1B6GH45 A0A0A9WLQ0 A0A0N0U4Y9 A0A1S3DDQ4 A0A0J7K5K3 A0A026WWI1 E2AMV8 A0A2P8Z694 A0A0L0BL52 T1D5H0 A0A0B6Z186 A0A034WJC3 A0A0C9SA70 A0A1E1WXD1 L7M2G9 W8BTT0 A0A0P6IUS3 A0A023FG34 A0A023FTF8 A0A1S3GLL4 A0A1I8Q1W6 A0A1S3ABQ7 A0A1L8EEF7 A0A1I8N996 A0A0K8UQ58 T1E8T5 W5JV84 A0A2M3ZBV2 A0A2M4AP52 A0A182FCJ7 G3MLR4 A0A023GHM2 A0A131XCJ5 A0A224Z3I7 A0A131YQF7 U3FPK9 A0A023EEY4 A0A182JJE6 E9IX33 H9H1M0 F8RT80 A0A1Q3FXM5 B0XED5 A0A2Y9EA22 A0A1A6H5Z3 I3M8N9 A0A250Y928 H0WCU5 A0A091CL71 B2GV64 G3HCV5 G5BYY0 A0A2K6EDD2 A0A2K6K9E0 A0A384DSD7 G1PKY9 G3RC27 F1PKC8 G1RQA1 A0A2K5Y946 A0A0D9R2R2 A0A2U3V3P2 A0A3Q7NKN9 A0A340Y658 A0A2R8ZEE2 A0A0A0MVA8 A0A2K6Q285 A0A2Y9IFB2
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
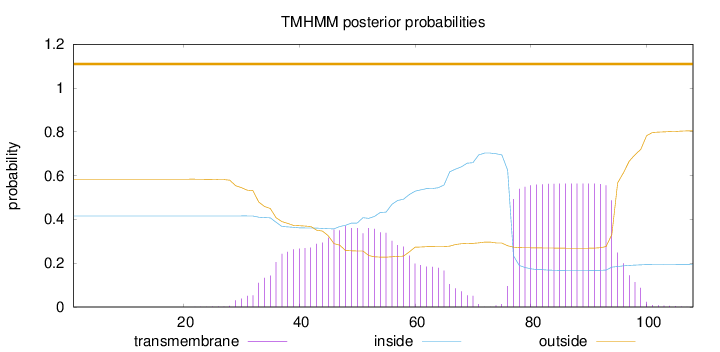
Length:
108
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
20.24163
Exp number, first 60 AAs:
8.06153
Total prob of N-in:
0.41617
outside
1 - 108
Population Genetic Test Statistics
Pi
140.697589
Theta
13.028017
Tajima's D
0.088708
CLR
6.850345
CSRT
0.3982800859957
Interpretation
Uncertain
