Gene
KWMTBOMO05798 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006937
Annotation
troponin_C_[Bombyx_mori]
Full name
Troponin C, isoform 3
Location in the cell
Cytoplasmic Reliability : 3.596
Sequence
CDS
ATGGTTCTGGACACCGAAGACTGGGCCGATGAGCTGCCCCCAGAACAGATCGCCGTCCTCCGCAAAGCCTTCGACGGCTTCGACCACAACCGCTCGGGTAGTATCCCCTGTGACTTCGTGGCTGATATCCTGAGAATGATGGGGCAGCCATTCAACAAGAAAATCCTCGAGGAACTTATTGAGGAGGTCGACGCCGACAAATCTGGTCGTCTGGAATTCCCCGAGTTCGTAACTCTGGCCGCTAAGTTCATCGTTGAAGAGGACGCTGAGGCCATGCAGAAAGAACTCAGGGAAGCGTTCAGATTGTACGACAAGGAAGGCAACGGTTACATTCCCACGTCGAGCCTGCGCGAGATCCTCCGGGAATTGGACGAGCAGCTGACTGACGATGAACTCGATGGCCTCATCCAAGAGATCGACACTGACGGAAGCGGCACCGTCGACTTCGACGAGTTCATGGAGATGATGACAGGGGAATAA
Protein
MVLDTEDWADELPPEQIAVLRKAFDGFDHNRSGSIPCDFVADILRMMGQPFNKKILEELIEEVDADKSGRLEFPEFVTLAAKFIVEEDAEAMQKELREAFRLYDKEGNGYIPTSSLREILRELDEQLTDDELDGLIQEIDTDGSGTVDFDEFMEMMTGE
Summary
Similarity
Belongs to the troponin C family.
Keywords
Calcium
Complete proteome
Metal-binding
Muscle protein
Reference proteome
Repeat
Feature
chain Troponin C, isoform 3
Uniprot
I4DLV8
Q5MGI5
C0LZ18
Q1HPK1
I4DIE2
S4PQ02
+ More
E7EC48 A0A3S2NMM9 A0A2H1V4I6 A0A212FF47 L0N756 A0A194QR58 A0A2W1C0C1 A0A2A4J1H3 K7J1T0 A0A088A1V5 V5IAE3 E0VPZ8 A0A158NTJ0 A0A1D1ZDP5 D6WZP8 K7J1S9 Q868D2 A0A0C9R6B0 A0A0C9QNU6 A0A170Y0S2 V5JDG8 A0A2R7WIY2 A0A224Y0G7 A0A0P4VRE5 A0A023F9W6 A0A069DW45 R4FPJ7 J3JVB0 A0A146M6Z2 A0A0A9WDW2 A0A146LDJ3 A0A195EEV5 A0A0V0GBM2 V5JDG4 R4WDQ8 A0A2H8TXM9 C4WYG9 A0A1Y1KPA5 A0A146M9Z3 A2I491 A0A1B6LW27 A0A1B6GKM8 A0A1B6DGE2 A0A2S2R557 A0A1W4XLE8 A0A0T6AYJ1 U4TYD9 H9JBP2 A0A1B0CDR0 B3M5H7 B4NPG0 A0A1L8DM14 B5DQG0 B4HBW5 I7LST2 B4KRQ0 B4IZD6 A0A0K8TY42 A0A034VRI4 W8BZR5 B4J8I0 B3MET4 A0A1W4W4N1 X2JCI6 B4PJY5 A0A0M4ECX9 B4HKA8 B3NDD4 B4QNE8 P47949 B4KYF0 B4LDT4 B4MFL8 B4MPC9 A0A0L0BTD6 D3TPS7 A0A1B0AG16 A0A1A9WFK2 A0A0J9RY91 A0A336KZQ2 A0A3Q0J911 R4UW10 W8C8Z7 Q6WEW7 T1PA43 W8BXG0 A0A2J7PW11 Q6WEW8 A0A3B0JM16 A0A1A9YG67 A0A1A9VCF8 Q1A7B1 A0A0A1X843 A0A0K8UE79
E7EC48 A0A3S2NMM9 A0A2H1V4I6 A0A212FF47 L0N756 A0A194QR58 A0A2W1C0C1 A0A2A4J1H3 K7J1T0 A0A088A1V5 V5IAE3 E0VPZ8 A0A158NTJ0 A0A1D1ZDP5 D6WZP8 K7J1S9 Q868D2 A0A0C9R6B0 A0A0C9QNU6 A0A170Y0S2 V5JDG8 A0A2R7WIY2 A0A224Y0G7 A0A0P4VRE5 A0A023F9W6 A0A069DW45 R4FPJ7 J3JVB0 A0A146M6Z2 A0A0A9WDW2 A0A146LDJ3 A0A195EEV5 A0A0V0GBM2 V5JDG4 R4WDQ8 A0A2H8TXM9 C4WYG9 A0A1Y1KPA5 A0A146M9Z3 A2I491 A0A1B6LW27 A0A1B6GKM8 A0A1B6DGE2 A0A2S2R557 A0A1W4XLE8 A0A0T6AYJ1 U4TYD9 H9JBP2 A0A1B0CDR0 B3M5H7 B4NPG0 A0A1L8DM14 B5DQG0 B4HBW5 I7LST2 B4KRQ0 B4IZD6 A0A0K8TY42 A0A034VRI4 W8BZR5 B4J8I0 B3MET4 A0A1W4W4N1 X2JCI6 B4PJY5 A0A0M4ECX9 B4HKA8 B3NDD4 B4QNE8 P47949 B4KYF0 B4LDT4 B4MFL8 B4MPC9 A0A0L0BTD6 D3TPS7 A0A1B0AG16 A0A1A9WFK2 A0A0J9RY91 A0A336KZQ2 A0A3Q0J911 R4UW10 W8C8Z7 Q6WEW7 T1PA43 W8BXG0 A0A2J7PW11 Q6WEW8 A0A3B0JM16 A0A1A9YG67 A0A1A9VCF8 Q1A7B1 A0A0A1X843 A0A0K8UE79
Pubmed
22651552
16023793
26354079
23622113
22118469
28756777
+ More
20075255 20566863 21347285 18362917 19820115 12558500 27538518 27129103 25474469 26334808 22516182 23537049 26823975 25401762 23691247 28004739 19121390 17994087 15632085 15696366 25348373 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 7980384 26108605 20353571 25315136 16751002 25830018
20075255 20566863 21347285 18362917 19820115 12558500 27538518 27129103 25474469 26334808 22516182 23537049 26823975 25401762 23691247 28004739 19121390 17994087 15632085 15696366 25348373 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 7980384 26108605 20353571 25315136 16751002 25830018
EMBL
AK402276
BAM18898.1
AY829801
AAV91415.1
FJ785831
ACN86371.1
+ More
DQ443401 DQ675020 ABF51490.1 ABG02550.1 AK401060 KQ459302 BAM17682.1 KPJ01781.1 GAIX01002120 JAA90440.1 HQ649758 ADV36664.1 RSAL01000003 RVE54857.1 ODYU01000639 SOQ35755.1 AGBW02008863 OWR52348.1 AK289350 BAM73876.1 KQ461175 KPJ07834.1 KZ149900 PZC78560.1 NWSH01003996 PCG65586.1 GALX01001335 JAB67131.1 DS235387 EEB15454.1 ADTU01025973 GDJX01002916 JAT65020.1 KQ971372 EFA10458.1 AJ512941 CAD55597.1 GBYB01002281 JAG72048.1 GBYB01002282 JAG72049.1 GEMB01003825 JAR99429.1 KC122901 KU932397 AGI96979.1 APA34033.1 KK854743 PTY18355.1 GFTR01002537 JAW13889.1 GDKW01000978 JAI55617.1 GBBI01000522 JAC18190.1 GBGD01003345 JAC85544.1 ACPB03005592 GAHY01000844 JAA76666.1 APGK01044268 APGK01044269 APGK01044270 APGK01044271 APGK01044272 BT127177 KB741022 AEE62139.1 ENN74977.1 GDHC01003088 JAQ15541.1 GBHO01037615 JAG05989.1 GDHC01013054 JAQ05575.1 KQ978983 KYN26805.1 GECL01001383 JAP04741.1 KC122902 AGI96980.1 AK417893 BAN21108.1 GFXV01006477 MBW18282.1 ABLF02023537 AK343182 BAH72939.1 GEZM01077591 JAV63259.1 GDHC01002048 JAQ16581.1 EF070594 ABM55660.1 GEBQ01012099 JAT27878.1 GECZ01006767 JAS63002.1 GEDC01012586 JAS24712.1 GGMS01015932 MBY85135.1 LJIG01022534 KRT80068.1 KB631843 ERL86654.1 BABH01024458 AJWK01008144 CH902618 EDV39587.1 CH964291 EDW86400.1 GFDF01006571 JAV07513.1 CH379069 EDY73882.1 CH479267 EDW39559.1 BK001457 DAA01505.1 CH933808 EDW08320.1 CH916366 EDV96691.1 GDHF01033314 JAI19000.1 GAKP01014794 JAC44158.1 GAMC01001688 JAC04868.1 CH916367 EDW02339.1 CH902619 EDV35548.1 AE014296 AHN58116.1 CM000159 EDW94754.1 CP012525 ALC43358.1 CH480815 EDW41849.1 CH954178 EDV51996.1 CM000363 CM002912 EDX10818.1 KMZ00189.1 X76042 BT099811 CH933809 EDW17729.1 CH940647 EDW68957.1 CH940667 EDW57189.1 CH963849 EDW73968.1 JRES01001371 KNC23307.1 EZ423429 ADD19705.1 KMZ00190.1 UFQS01000862 UFQS01001231 UFQT01000862 UFQT01001231 SSX07349.1 SSX09866.1 SSX27691.1 KC571907 AGM32406.1 GAMC01002609 JAC03947.1 AY292254 AAQ57577.1 KA645559 AFP60188.1 GAMC01002608 JAC03948.1 NEVH01020940 PNF20521.1 AY292253 AAQ57576.1 OUUW01000001 SPP74589.1 DQ279094 ABB89298.1 GBXI01007392 JAD06900.1 GDHF01027446 GDHF01001820 JAI24868.1 JAI50494.1
DQ443401 DQ675020 ABF51490.1 ABG02550.1 AK401060 KQ459302 BAM17682.1 KPJ01781.1 GAIX01002120 JAA90440.1 HQ649758 ADV36664.1 RSAL01000003 RVE54857.1 ODYU01000639 SOQ35755.1 AGBW02008863 OWR52348.1 AK289350 BAM73876.1 KQ461175 KPJ07834.1 KZ149900 PZC78560.1 NWSH01003996 PCG65586.1 GALX01001335 JAB67131.1 DS235387 EEB15454.1 ADTU01025973 GDJX01002916 JAT65020.1 KQ971372 EFA10458.1 AJ512941 CAD55597.1 GBYB01002281 JAG72048.1 GBYB01002282 JAG72049.1 GEMB01003825 JAR99429.1 KC122901 KU932397 AGI96979.1 APA34033.1 KK854743 PTY18355.1 GFTR01002537 JAW13889.1 GDKW01000978 JAI55617.1 GBBI01000522 JAC18190.1 GBGD01003345 JAC85544.1 ACPB03005592 GAHY01000844 JAA76666.1 APGK01044268 APGK01044269 APGK01044270 APGK01044271 APGK01044272 BT127177 KB741022 AEE62139.1 ENN74977.1 GDHC01003088 JAQ15541.1 GBHO01037615 JAG05989.1 GDHC01013054 JAQ05575.1 KQ978983 KYN26805.1 GECL01001383 JAP04741.1 KC122902 AGI96980.1 AK417893 BAN21108.1 GFXV01006477 MBW18282.1 ABLF02023537 AK343182 BAH72939.1 GEZM01077591 JAV63259.1 GDHC01002048 JAQ16581.1 EF070594 ABM55660.1 GEBQ01012099 JAT27878.1 GECZ01006767 JAS63002.1 GEDC01012586 JAS24712.1 GGMS01015932 MBY85135.1 LJIG01022534 KRT80068.1 KB631843 ERL86654.1 BABH01024458 AJWK01008144 CH902618 EDV39587.1 CH964291 EDW86400.1 GFDF01006571 JAV07513.1 CH379069 EDY73882.1 CH479267 EDW39559.1 BK001457 DAA01505.1 CH933808 EDW08320.1 CH916366 EDV96691.1 GDHF01033314 JAI19000.1 GAKP01014794 JAC44158.1 GAMC01001688 JAC04868.1 CH916367 EDW02339.1 CH902619 EDV35548.1 AE014296 AHN58116.1 CM000159 EDW94754.1 CP012525 ALC43358.1 CH480815 EDW41849.1 CH954178 EDV51996.1 CM000363 CM002912 EDX10818.1 KMZ00189.1 X76042 BT099811 CH933809 EDW17729.1 CH940647 EDW68957.1 CH940667 EDW57189.1 CH963849 EDW73968.1 JRES01001371 KNC23307.1 EZ423429 ADD19705.1 KMZ00190.1 UFQS01000862 UFQS01001231 UFQT01000862 UFQT01001231 SSX07349.1 SSX09866.1 SSX27691.1 KC571907 AGM32406.1 GAMC01002609 JAC03947.1 AY292254 AAQ57577.1 KA645559 AFP60188.1 GAMC01002608 JAC03948.1 NEVH01020940 PNF20521.1 AY292253 AAQ57576.1 OUUW01000001 SPP74589.1 DQ279094 ABB89298.1 GBXI01007392 JAD06900.1 GDHF01027446 GDHF01001820 JAI24868.1 JAI50494.1
Proteomes
UP000053268
UP000283053
UP000007151
UP000053240
UP000218220
UP000002358
+ More
UP000005203 UP000009046 UP000005205 UP000007266 UP000015103 UP000019118 UP000078492 UP000007819 UP000192223 UP000030742 UP000005204 UP000092461 UP000007801 UP000007798 UP000001819 UP000008744 UP000009192 UP000001070 UP000192221 UP000000803 UP000002282 UP000092553 UP000001292 UP000008711 UP000000304 UP000008792 UP000037069 UP000092445 UP000091820 UP000079169 UP000095301 UP000235965 UP000268350 UP000092443 UP000078200
UP000005203 UP000009046 UP000005205 UP000007266 UP000015103 UP000019118 UP000078492 UP000007819 UP000192223 UP000030742 UP000005204 UP000092461 UP000007801 UP000007798 UP000001819 UP000008744 UP000009192 UP000001070 UP000192221 UP000000803 UP000002282 UP000092553 UP000001292 UP000008711 UP000000304 UP000008792 UP000037069 UP000092445 UP000091820 UP000079169 UP000095301 UP000235965 UP000268350 UP000092443 UP000078200
SUPFAM
SSF47473
SSF47473
CDD
ProteinModelPortal
I4DLV8
Q5MGI5
C0LZ18
Q1HPK1
I4DIE2
S4PQ02
+ More
E7EC48 A0A3S2NMM9 A0A2H1V4I6 A0A212FF47 L0N756 A0A194QR58 A0A2W1C0C1 A0A2A4J1H3 K7J1T0 A0A088A1V5 V5IAE3 E0VPZ8 A0A158NTJ0 A0A1D1ZDP5 D6WZP8 K7J1S9 Q868D2 A0A0C9R6B0 A0A0C9QNU6 A0A170Y0S2 V5JDG8 A0A2R7WIY2 A0A224Y0G7 A0A0P4VRE5 A0A023F9W6 A0A069DW45 R4FPJ7 J3JVB0 A0A146M6Z2 A0A0A9WDW2 A0A146LDJ3 A0A195EEV5 A0A0V0GBM2 V5JDG4 R4WDQ8 A0A2H8TXM9 C4WYG9 A0A1Y1KPA5 A0A146M9Z3 A2I491 A0A1B6LW27 A0A1B6GKM8 A0A1B6DGE2 A0A2S2R557 A0A1W4XLE8 A0A0T6AYJ1 U4TYD9 H9JBP2 A0A1B0CDR0 B3M5H7 B4NPG0 A0A1L8DM14 B5DQG0 B4HBW5 I7LST2 B4KRQ0 B4IZD6 A0A0K8TY42 A0A034VRI4 W8BZR5 B4J8I0 B3MET4 A0A1W4W4N1 X2JCI6 B4PJY5 A0A0M4ECX9 B4HKA8 B3NDD4 B4QNE8 P47949 B4KYF0 B4LDT4 B4MFL8 B4MPC9 A0A0L0BTD6 D3TPS7 A0A1B0AG16 A0A1A9WFK2 A0A0J9RY91 A0A336KZQ2 A0A3Q0J911 R4UW10 W8C8Z7 Q6WEW7 T1PA43 W8BXG0 A0A2J7PW11 Q6WEW8 A0A3B0JM16 A0A1A9YG67 A0A1A9VCF8 Q1A7B1 A0A0A1X843 A0A0K8UE79
E7EC48 A0A3S2NMM9 A0A2H1V4I6 A0A212FF47 L0N756 A0A194QR58 A0A2W1C0C1 A0A2A4J1H3 K7J1T0 A0A088A1V5 V5IAE3 E0VPZ8 A0A158NTJ0 A0A1D1ZDP5 D6WZP8 K7J1S9 Q868D2 A0A0C9R6B0 A0A0C9QNU6 A0A170Y0S2 V5JDG8 A0A2R7WIY2 A0A224Y0G7 A0A0P4VRE5 A0A023F9W6 A0A069DW45 R4FPJ7 J3JVB0 A0A146M6Z2 A0A0A9WDW2 A0A146LDJ3 A0A195EEV5 A0A0V0GBM2 V5JDG4 R4WDQ8 A0A2H8TXM9 C4WYG9 A0A1Y1KPA5 A0A146M9Z3 A2I491 A0A1B6LW27 A0A1B6GKM8 A0A1B6DGE2 A0A2S2R557 A0A1W4XLE8 A0A0T6AYJ1 U4TYD9 H9JBP2 A0A1B0CDR0 B3M5H7 B4NPG0 A0A1L8DM14 B5DQG0 B4HBW5 I7LST2 B4KRQ0 B4IZD6 A0A0K8TY42 A0A034VRI4 W8BZR5 B4J8I0 B3MET4 A0A1W4W4N1 X2JCI6 B4PJY5 A0A0M4ECX9 B4HKA8 B3NDD4 B4QNE8 P47949 B4KYF0 B4LDT4 B4MFL8 B4MPC9 A0A0L0BTD6 D3TPS7 A0A1B0AG16 A0A1A9WFK2 A0A0J9RY91 A0A336KZQ2 A0A3Q0J911 R4UW10 W8C8Z7 Q6WEW7 T1PA43 W8BXG0 A0A2J7PW11 Q6WEW8 A0A3B0JM16 A0A1A9YG67 A0A1A9VCF8 Q1A7B1 A0A0A1X843 A0A0K8UE79
PDB
4IL1
E-value=5.1637e-26,
Score=287
Ontologies
GO
Topology
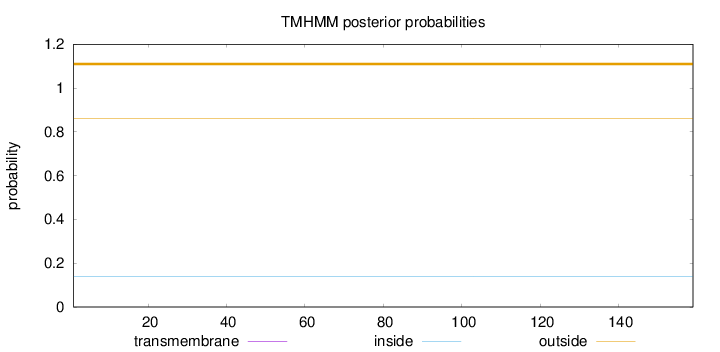
Length:
159
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00019
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.13990
outside
1 - 159
Population Genetic Test Statistics
Pi
192.326042
Theta
163.913746
Tajima's D
0.360817
CLR
0.288157
CSRT
0.466226688665567
Interpretation
Uncertain
