Gene
KWMTBOMO05794
Pre Gene Modal
BGIBMGA006936
Annotation
cytochrome_P450_[Bombyx_mori]
Full name
Cytochrome P450 306a1
Alternative Name
Protein phantom
Location in the cell
Mitochondrial Reliability : 1.57 PlasmaMembrane Reliability : 1.502
Sequence
CDS
ATGGACCTTTATTTTATTTGGCTGGTAACGTTCGTGGCTGGGTTTTGGATTTTCAAAAAAATAAAGGAATGGCAGAATTTGCCCCCCGGACCTTGGGGGTTACCTATCGTCGGTTATTTGCCTTTCATTGATCGCTATCATCCACATATCACCTTGACAAATTTGTCTAAAACATACGGAGCTATTTACGGTCTCAAAATGGGCAGCATATATGCTGTAGTGTTATCTGATCATAAACTTGTAAGAGATACGTTCTCAAAAGACAGTTTTTCTGGACGAGCACCTCTTTACTTAACACATGGCCTTATGAATGGAAATGGAATTATTTGTGCCGAAGGCGGTTTGTGGAGGGACCAAAGGAAATTAATAACATCGTGGTTGAAAAGTTTTGGAATGAGTAAGCACAGTGTTTCCCGAGAAAAATTGGAAAAACGAATCGCTTCAGGAGTATACGAAATTTTGGAAAATATCGAAAAAACTTCTGATGCTGCCTTGGACCTTCCTCATATGCTGACGAATTCTTTAGGAAACGTTGTCAATGAGATAATATTCGGCTTTAAGTTTCCACCTGAAAATAAAACATGGCAATGGTTTCGTCAAATACAGGAGGAAGGATGCCATGAGATGGGAGTCGCAGGTGTTGTAAACTTCTTGCCCTTTATACGCCATGTTTCGCCATCAACACGAAAAACAATTGAAGTTCTAGTCCGTGGACAGGCACAGACGCATACTTTGTACGCAAGCATGATAGATAGACGAAGAAAAATGTTGGGCTTAGAGAAGCCTAAGGGAGCCGAATATGCTCCTCACGAAAACCTTTTAAAACTATATCCAAATGGCCATATCAAATGCATAAAATACAGCAAAGTCTCTCCGAACACCGAGCATTTCTTTGATCCCAATACTCTTATTCCGACTGAAGGAGATTGCATACTGGATAATTTTCTGTTGGAGCAAAAGAAACGATTTGAGAGTGGAGACCCAACGGCACTGTATATGAGAGATGAACAGTTACATTTTCTACTAGCGGATATGTTCGGTGCCGGACTGGATACTACATCGGTGACTTTAGCTTGGTTTTTGCTATACATGGCTTTGTTTCCAGAAGAACAGGAAGAAATACGTAAAGAAATCTTATCCGTATATCCATATGACGATGATGTTGATAGTTCAAGGTTACCTCTTCTTATGGCAGCAATCTGTGAAACTCAGAGGATTCGATCGATTGTTCCAGTGGGAATACCCCATGGTTGTATAGAGGACGCTTACTTGGGTAACTACAGAATCCCAAAAAATGCCATGGTGATCCCATTGCAGTGGGCTATTCACATGGATCCTAATGTTTGGGAAGAACCAGAAAAATTCAAACCGCGTAGATTTTTGGCTCAGGATGGTAGTCTACTTAAGCCTCAAGAATTCATTCCGTTTCAAACTGGTAAGCGGATGTGTCCGGGTGACGAACTGTCCCGTATGTTGTCGTGTGGCCTCGTAAGTAGACTATTCAGAAAGCAGCGTATTCGACTCGCATCAAAAATACCGACAGCAGAAGAGATGCGTGGAACCGTCGGTGTTACGTTGGCACCTCCTCCGGTGAAATACTATTGCGAACCAATTTAA
Protein
MDLYFIWLVTFVAGFWIFKKIKEWQNLPPGPWGLPIVGYLPFIDRYHPHITLTNLSKTYGAIYGLKMGSIYAVVLSDHKLVRDTFSKDSFSGRAPLYLTHGLMNGNGIICAEGGLWRDQRKLITSWLKSFGMSKHSVSREKLEKRIASGVYEILENIEKTSDAALDLPHMLTNSLGNVVNEIIFGFKFPPENKTWQWFRQIQEEGCHEMGVAGVVNFLPFIRHVSPSTRKTIEVLVRGQAQTHTLYASMIDRRRKMLGLEKPKGAEYAPHENLLKLYPNGHIKCIKYSKVSPNTEHFFDPNTLIPTEGDCILDNFLLEQKKRFESGDPTALYMRDEQLHFLLADMFGAGLDTTSVTLAWFLLYMALFPEEQEEIRKEILSVYPYDDDVDSSRLPLLMAAICETQRIRSIVPVGIPHGCIEDAYLGNYRIPKNAMVIPLQWAIHMDPNVWEEPEKFKPRRFLAQDGSLLKPQEFIPFQTGKRMCPGDELSRMLSCGLVSRLFRKQRIRLASKIPTAEEMRGTVGVTLAPPPVKYYCEPI
Summary
Description
Involved in the metabolism of insect hormones; responsible for ecdysteroid C25-hydroxylase activity. May be involved in the breakdown of synthetic insecticides.
Cofactor
heme
Miscellaneous
Member of the Halloween gene group.
Similarity
Belongs to the cytochrome P450 family.
Keywords
Complete proteome
Endoplasmic reticulum
Heme
Iron
Membrane
Metal-binding
Microsome
Monooxygenase
Oxidoreductase
Polymorphism
Reference proteome
Feature
chain Cytochrome P450 306a1
Uniprot
H9JBP1
Q6BDY0
L0N5I6
Q6I6X0
Q2HZZ6
A0A2W1C0A8
+ More
A0A068F0T6 A0A2A4JAS3 D1FQH3 A0A248QEY9 A0A0P0I325 A0A1E1WND1 A0A1E1W181 A0A1E1WGU8 S6BAP6 A0A0C5C1I0 A0A194QS71 A0A1V0D9F2 A0A212FF22 X5D9A7 A0A194Q8R4 A0A286MXN3 A0A0L7LQ90 W5JAY4 Q17BQ0 A0A182NN31 A0A158ND44 A0A2P8XVJ5 A0A067RMS6 A0A182F8T9 A0A088ASS9 A0A195BM36 A0A336MQD5 K7JC63 A0A232F297 A0A182YEM7 A0A0L7R6V6 A0A1A9X5F1 A9Q615 Q7Q2D4 A0A195F7N7 A0A182RKU0 A0A0N0U373 V5K5D2 W8B5H1 B3MVQ6 A0A182VPZ7 A0A0A1XKL1 A0A0L0CI01 D6X1E7 A0A195DUI8 A0A034W432 A0A3L8E518 A0A0K8V4M9 A0A182PDH3 A0A0K0LB83 A0A3B0JR04 E0VMV6 A0A310S4V1 B4ND96 B4L7K0 B4PWD8 X2JG03 Q9VWR5 A0A0C9RAE0 A0A1I8N403 B4JJR1 M9SXQ0 A0A1Q1NKZ3 B3NUM7 A0A0M4HXV3 B4M2S7 A0A0A9YSL0 A0A1W4UDL8 A0A1B6K405 M9T1D3 B9X0J8 K4JNB8 A0A1J1J5W8 A0A2A3EB12 A0A182KDN2 A0A0P5U6B1 A0A0U1W0T7 A0A0P5T266 A0A2R7WE89 A0A224XII9 A0A0P5I5K7 A0A0P5N6P9 A0A0P5HTM4 A0A0P6BPE0 A0A0P6CW43 A0A069DUN2 A0A0N8C1E8 A0A0P5G153 A0A0P6I8Y2 A0A0P5PV74 A0A0P5JWR0 A0A0N8BTG0 A0A0P5PXS3
A0A068F0T6 A0A2A4JAS3 D1FQH3 A0A248QEY9 A0A0P0I325 A0A1E1WND1 A0A1E1W181 A0A1E1WGU8 S6BAP6 A0A0C5C1I0 A0A194QS71 A0A1V0D9F2 A0A212FF22 X5D9A7 A0A194Q8R4 A0A286MXN3 A0A0L7LQ90 W5JAY4 Q17BQ0 A0A182NN31 A0A158ND44 A0A2P8XVJ5 A0A067RMS6 A0A182F8T9 A0A088ASS9 A0A195BM36 A0A336MQD5 K7JC63 A0A232F297 A0A182YEM7 A0A0L7R6V6 A0A1A9X5F1 A9Q615 Q7Q2D4 A0A195F7N7 A0A182RKU0 A0A0N0U373 V5K5D2 W8B5H1 B3MVQ6 A0A182VPZ7 A0A0A1XKL1 A0A0L0CI01 D6X1E7 A0A195DUI8 A0A034W432 A0A3L8E518 A0A0K8V4M9 A0A182PDH3 A0A0K0LB83 A0A3B0JR04 E0VMV6 A0A310S4V1 B4ND96 B4L7K0 B4PWD8 X2JG03 Q9VWR5 A0A0C9RAE0 A0A1I8N403 B4JJR1 M9SXQ0 A0A1Q1NKZ3 B3NUM7 A0A0M4HXV3 B4M2S7 A0A0A9YSL0 A0A1W4UDL8 A0A1B6K405 M9T1D3 B9X0J8 K4JNB8 A0A1J1J5W8 A0A2A3EB12 A0A182KDN2 A0A0P5U6B1 A0A0U1W0T7 A0A0P5T266 A0A2R7WE89 A0A224XII9 A0A0P5I5K7 A0A0P5N6P9 A0A0P5HTM4 A0A0P6BPE0 A0A0P6CW43 A0A069DUN2 A0A0N8C1E8 A0A0P5G153 A0A0P6I8Y2 A0A0P5PV74 A0A0P5JWR0 A0A0N8BTG0 A0A0P5PXS3
EC Number
1.14.-.-
Pubmed
19121390
15350618
15197185
16503480
28756777
19682519
+ More
28881445 26354079 22118469 24361403 19954531 25299618 26227816 20920257 23761445 17510324 21347285 29403074 24845553 20075255 28648823 25244985 19060216 12364791 14747013 17210077 24267698 24495485 17994087 25830018 26108605 18362917 19820115 25348373 30249741 20566863 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17322555 12537569 25315136 28092127 26319543 25401762 26823975 25017052 24324548 26334808
28881445 26354079 22118469 24361403 19954531 25299618 26227816 20920257 23761445 17510324 21347285 29403074 24845553 20075255 28648823 25244985 19060216 12364791 14747013 17210077 24267698 24495485 17994087 25830018 26108605 18362917 19820115 25348373 30249741 20566863 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17322555 12537569 25315136 28092127 26319543 25401762 26823975 25017052 24324548 26334808
EMBL
BABH01024447
AK289327
BAM73854.1
AB124840
BAD34476.1
AK289326
+ More
BAM73853.1 AB162964 BAD23844.1 DQ357066 ABC96068.1 KZ149900 PZC78550.1 KM016703 AID54855.1 NWSH01002327 PCG68520.1 FJ010194 ACM45975.1 KX443455 ASO98030.1 KP764854 ALJ84052.1 GDQN01002521 JAT88533.1 GDQN01010294 JAT80760.1 GDQN01004845 JAT86209.1 AB649117 BAN66311.1 KP001121 AJN91166.1 KQ461175 KPJ07830.1 KY212066 ARA91630.1 AGBW02008863 OWR52346.1 KF701127 AHW57297.1 KQ459302 KPJ01784.1 MF684351 ASX93985.1 JTDY01000352 KOB77579.1 ADMH02002005 ETN60005.1 CH477318 EAT43717.2 ADTU01012323 ADTU01012324 ADTU01012325 PYGN01001282 PSN36025.1 KK852530 KDR21920.1 KQ976438 KYM86821.1 UFQS01000959 UFQT01000959 SSX07954.1 SSX28188.1 AAZX01000906 NNAY01001255 OXU24598.1 KQ414646 KOC66605.1 EF546762 ABU42522.1 AAAB01008977 EAA13571.4 KQ981744 KYN36453.1 KQ435936 KOX68308.1 KF044261 AGT57833.1 GAMC01010170 JAB96385.1 CH902625 EDV35051.2 GBXI01002877 JAD11415.1 JRES01000348 KNC32033.1 KQ971368 EFA10665.2 KQ980341 KYN16422.1 GAKP01008621 JAC50331.1 QOIP01000001 RLU27513.1 GDHF01018493 JAI33821.1 KM217013 AIW79976.1 OUUW01000003 SPP77900.1 DS235327 EEB14712.1 KQ774645 OAD52305.1 CH964239 EDW82805.1 CH933814 EDW05713.1 KRG07498.1 CM000162 EDX02756.1 AE014298 AHN59884.1 AF484413 AM411848 AM411849 AM411850 AM411851 AM411852 AM411853 AM411854 AM411855 AM411856 AM411857 AM411858 AM411859 AM411860 AY070930 GBYB01013414 JAG83181.1 CH916370 EDV99813.1 KC701465 KC579463 AGI92300.1 AGU16452.1 KX660714 AQM57053.1 CH954180 EDV46624.1 KR012857 ALD15932.1 CH940651 EDW65981.1 GBHO01008436 GBHO01008435 GDHC01020222 GDHC01015501 JAG35168.1 JAG35169.1 JAP98406.1 JAQ03128.1 GECU01007129 GECU01001531 JAT00578.1 JAT06176.1 KC701463 KC579458 AGI92298.1 AGU16447.1 AB455969 BAH24005.1 JX566820 AFU86481.1 CVRI01000073 CRL07789.1 KZ288312 PBC28376.1 GDIP01119248 JAL84466.1 KC688866 AGT79088.1 GDIP01134441 JAL69273.1 KK854638 PTY17541.1 GFTR01006788 JAW09638.1 GDIQ01223097 JAK28628.1 GDIQ01146578 JAL05148.1 GDIQ01223098 JAK28627.1 GDIP01013565 JAM90150.1 GDIQ01087694 JAN07043.1 GBGD01001234 JAC87655.1 GDIQ01124275 JAL27451.1 GDIQ01252696 JAJ99028.1 GDIQ01033721 JAN61016.1 GDIQ01143824 JAL07902.1 GDIQ01198314 JAK53411.1 GDIQ01146579 GDIQ01146577 JAL05147.1 GDIQ01121930 JAL29796.1
BAM73853.1 AB162964 BAD23844.1 DQ357066 ABC96068.1 KZ149900 PZC78550.1 KM016703 AID54855.1 NWSH01002327 PCG68520.1 FJ010194 ACM45975.1 KX443455 ASO98030.1 KP764854 ALJ84052.1 GDQN01002521 JAT88533.1 GDQN01010294 JAT80760.1 GDQN01004845 JAT86209.1 AB649117 BAN66311.1 KP001121 AJN91166.1 KQ461175 KPJ07830.1 KY212066 ARA91630.1 AGBW02008863 OWR52346.1 KF701127 AHW57297.1 KQ459302 KPJ01784.1 MF684351 ASX93985.1 JTDY01000352 KOB77579.1 ADMH02002005 ETN60005.1 CH477318 EAT43717.2 ADTU01012323 ADTU01012324 ADTU01012325 PYGN01001282 PSN36025.1 KK852530 KDR21920.1 KQ976438 KYM86821.1 UFQS01000959 UFQT01000959 SSX07954.1 SSX28188.1 AAZX01000906 NNAY01001255 OXU24598.1 KQ414646 KOC66605.1 EF546762 ABU42522.1 AAAB01008977 EAA13571.4 KQ981744 KYN36453.1 KQ435936 KOX68308.1 KF044261 AGT57833.1 GAMC01010170 JAB96385.1 CH902625 EDV35051.2 GBXI01002877 JAD11415.1 JRES01000348 KNC32033.1 KQ971368 EFA10665.2 KQ980341 KYN16422.1 GAKP01008621 JAC50331.1 QOIP01000001 RLU27513.1 GDHF01018493 JAI33821.1 KM217013 AIW79976.1 OUUW01000003 SPP77900.1 DS235327 EEB14712.1 KQ774645 OAD52305.1 CH964239 EDW82805.1 CH933814 EDW05713.1 KRG07498.1 CM000162 EDX02756.1 AE014298 AHN59884.1 AF484413 AM411848 AM411849 AM411850 AM411851 AM411852 AM411853 AM411854 AM411855 AM411856 AM411857 AM411858 AM411859 AM411860 AY070930 GBYB01013414 JAG83181.1 CH916370 EDV99813.1 KC701465 KC579463 AGI92300.1 AGU16452.1 KX660714 AQM57053.1 CH954180 EDV46624.1 KR012857 ALD15932.1 CH940651 EDW65981.1 GBHO01008436 GBHO01008435 GDHC01020222 GDHC01015501 JAG35168.1 JAG35169.1 JAP98406.1 JAQ03128.1 GECU01007129 GECU01001531 JAT00578.1 JAT06176.1 KC701463 KC579458 AGI92298.1 AGU16447.1 AB455969 BAH24005.1 JX566820 AFU86481.1 CVRI01000073 CRL07789.1 KZ288312 PBC28376.1 GDIP01119248 JAL84466.1 KC688866 AGT79088.1 GDIP01134441 JAL69273.1 KK854638 PTY17541.1 GFTR01006788 JAW09638.1 GDIQ01223097 JAK28628.1 GDIQ01146578 JAL05148.1 GDIQ01223098 JAK28627.1 GDIP01013565 JAM90150.1 GDIQ01087694 JAN07043.1 GBGD01001234 JAC87655.1 GDIQ01124275 JAL27451.1 GDIQ01252696 JAJ99028.1 GDIQ01033721 JAN61016.1 GDIQ01143824 JAL07902.1 GDIQ01198314 JAK53411.1 GDIQ01146579 GDIQ01146577 JAL05147.1 GDIQ01121930 JAL29796.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000000673 UP000008820 UP000075884 UP000005205 UP000245037 UP000027135 UP000069272 UP000005203 UP000078540 UP000002358 UP000215335 UP000076408 UP000053825 UP000091820 UP000007062 UP000078541 UP000075900 UP000053105 UP000007801 UP000075920 UP000037069 UP000007266 UP000078492 UP000279307 UP000075885 UP000268350 UP000009046 UP000007798 UP000009192 UP000002282 UP000000803 UP000095301 UP000001070 UP000008711 UP000008792 UP000192221 UP000183832 UP000242457 UP000075881
UP000000673 UP000008820 UP000075884 UP000005205 UP000245037 UP000027135 UP000069272 UP000005203 UP000078540 UP000002358 UP000215335 UP000076408 UP000053825 UP000091820 UP000007062 UP000078541 UP000075900 UP000053105 UP000007801 UP000075920 UP000037069 UP000007266 UP000078492 UP000279307 UP000075885 UP000268350 UP000009046 UP000007798 UP000009192 UP000002282 UP000000803 UP000095301 UP000001070 UP000008711 UP000008792 UP000192221 UP000183832 UP000242457 UP000075881
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
H9JBP1
Q6BDY0
L0N5I6
Q6I6X0
Q2HZZ6
A0A2W1C0A8
+ More
A0A068F0T6 A0A2A4JAS3 D1FQH3 A0A248QEY9 A0A0P0I325 A0A1E1WND1 A0A1E1W181 A0A1E1WGU8 S6BAP6 A0A0C5C1I0 A0A194QS71 A0A1V0D9F2 A0A212FF22 X5D9A7 A0A194Q8R4 A0A286MXN3 A0A0L7LQ90 W5JAY4 Q17BQ0 A0A182NN31 A0A158ND44 A0A2P8XVJ5 A0A067RMS6 A0A182F8T9 A0A088ASS9 A0A195BM36 A0A336MQD5 K7JC63 A0A232F297 A0A182YEM7 A0A0L7R6V6 A0A1A9X5F1 A9Q615 Q7Q2D4 A0A195F7N7 A0A182RKU0 A0A0N0U373 V5K5D2 W8B5H1 B3MVQ6 A0A182VPZ7 A0A0A1XKL1 A0A0L0CI01 D6X1E7 A0A195DUI8 A0A034W432 A0A3L8E518 A0A0K8V4M9 A0A182PDH3 A0A0K0LB83 A0A3B0JR04 E0VMV6 A0A310S4V1 B4ND96 B4L7K0 B4PWD8 X2JG03 Q9VWR5 A0A0C9RAE0 A0A1I8N403 B4JJR1 M9SXQ0 A0A1Q1NKZ3 B3NUM7 A0A0M4HXV3 B4M2S7 A0A0A9YSL0 A0A1W4UDL8 A0A1B6K405 M9T1D3 B9X0J8 K4JNB8 A0A1J1J5W8 A0A2A3EB12 A0A182KDN2 A0A0P5U6B1 A0A0U1W0T7 A0A0P5T266 A0A2R7WE89 A0A224XII9 A0A0P5I5K7 A0A0P5N6P9 A0A0P5HTM4 A0A0P6BPE0 A0A0P6CW43 A0A069DUN2 A0A0N8C1E8 A0A0P5G153 A0A0P6I8Y2 A0A0P5PV74 A0A0P5JWR0 A0A0N8BTG0 A0A0P5PXS3
A0A068F0T6 A0A2A4JAS3 D1FQH3 A0A248QEY9 A0A0P0I325 A0A1E1WND1 A0A1E1W181 A0A1E1WGU8 S6BAP6 A0A0C5C1I0 A0A194QS71 A0A1V0D9F2 A0A212FF22 X5D9A7 A0A194Q8R4 A0A286MXN3 A0A0L7LQ90 W5JAY4 Q17BQ0 A0A182NN31 A0A158ND44 A0A2P8XVJ5 A0A067RMS6 A0A182F8T9 A0A088ASS9 A0A195BM36 A0A336MQD5 K7JC63 A0A232F297 A0A182YEM7 A0A0L7R6V6 A0A1A9X5F1 A9Q615 Q7Q2D4 A0A195F7N7 A0A182RKU0 A0A0N0U373 V5K5D2 W8B5H1 B3MVQ6 A0A182VPZ7 A0A0A1XKL1 A0A0L0CI01 D6X1E7 A0A195DUI8 A0A034W432 A0A3L8E518 A0A0K8V4M9 A0A182PDH3 A0A0K0LB83 A0A3B0JR04 E0VMV6 A0A310S4V1 B4ND96 B4L7K0 B4PWD8 X2JG03 Q9VWR5 A0A0C9RAE0 A0A1I8N403 B4JJR1 M9SXQ0 A0A1Q1NKZ3 B3NUM7 A0A0M4HXV3 B4M2S7 A0A0A9YSL0 A0A1W4UDL8 A0A1B6K405 M9T1D3 B9X0J8 K4JNB8 A0A1J1J5W8 A0A2A3EB12 A0A182KDN2 A0A0P5U6B1 A0A0U1W0T7 A0A0P5T266 A0A2R7WE89 A0A224XII9 A0A0P5I5K7 A0A0P5N6P9 A0A0P5HTM4 A0A0P6BPE0 A0A0P6CW43 A0A069DUN2 A0A0N8C1E8 A0A0P5G153 A0A0P6I8Y2 A0A0P5PV74 A0A0P5JWR0 A0A0N8BTG0 A0A0P5PXS3
PDB
4R21
E-value=5.44173e-43,
Score=440
Ontologies
GO
GO:0005506
GO:0016705
GO:0020037
GO:0004497
GO:0016021
GO:0006082
GO:0005737
GO:0006805
GO:0016712
GO:0042738
GO:0043231
GO:0055114
GO:0008395
GO:0006697
GO:0007298
GO:0001700
GO:0035302
GO:0005783
GO:0070330
GO:0009307
GO:0003677
GO:0009036
GO:0048477
GO:0005789
GO:0051861
GO:0000160
GO:0005216
GO:0006811
GO:0006541
GO:0016812
GO:0016597
Topology
Subcellular location
Endoplasmic reticulum membrane
Microsome membrane
Microsome membrane
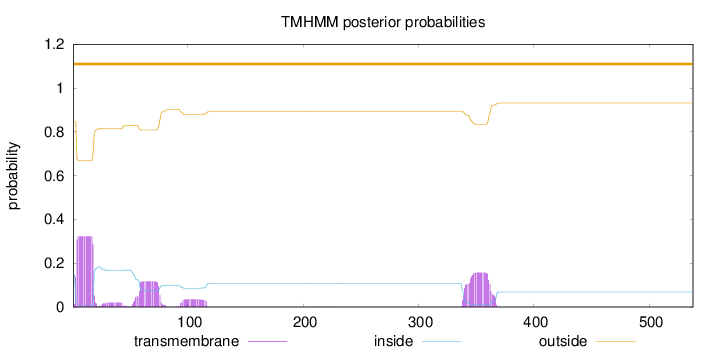
Length:
538
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.0553899999999
Exp number, first 60 AAs:
5.92028
Total prob of N-in:
0.15451
outside
1 - 538
Population Genetic Test Statistics
Pi
47.713086
Theta
54.854982
Tajima's D
0.437853
CLR
1.858675
CSRT
0.498125093745313
Interpretation
Uncertain
