Gene
KWMTBOMO05790
Pre Gene Modal
BGIBMGA006935
Annotation
PREDICTED:_transmembrane_protein_132B_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.407
Sequence
CDS
ATGGAACGGTGCCGGCTGGATGTTTCGGCTCATTTAGTCTTCCGAGAACTGCCCCGAGACTCGCCTGTACTACGTGTACTGTTTCACACAGGGGGGGAAGGTGCACGAAGAGCAGCATCTAGACCTCGACGTGTTTGCATAACATTGCATGCGAGTCTTGGATCGAAATCGTTAACGGCCACATGTGGTCCAGAAGGCGAAGAGGGGGCTTGCTTGGCTGAATTAACAGTTCCTGCAGCGTGGTGGCCAGCTGACGGTAAAGGAAAAAGACCACCAAGAACTGTTAGGTTAGCTTACAGTGCCACAGAAGCTGGCAATGGTGAATCCGGAGAGGATACGTGCGGTCGGGTCTCAGTACAACCAGCCTGGCCATTGGGATCCGTGATTCTTGCGGGTGCAAGAGCAGGTTATAGAGAAGCAAGAGCCGGCGATGCAGCCCTCCTTCTACCACGAGCACCTCTGTATCCGCATTCTAGAATACATTTACCTTTTGTTGTTCGTCGTGACAGTTCTCACAATATAGGACACGTTGCCATCAGAATGAAAGTCAAGTCGGGATTGAGGCTGCTAAGTGTTACGGCTGCCAACTCGCGCTGGGCTGTCACTTCAGAAGTAAAGATGCGCTCTGCGACTGTCACAGCTACAAGATTGGAGCAGCCACTGCGGGATGATGACGATATTGACTTTGATGAAGAAGAAACTATTGGAGCCGGAGGTCCTGGTTGGGAAGAAGTACTTACTTGGCTGGTGGAAGTTGGGCCTGAAGGTGAAGGTGACGCTGAGGGGTCCGAAGGCGAATCTGGAAGGGGTACAGCACGTTTGAGCTGGTCAGCCAAAGTTTCTCCAATAATATCTTCGTCTTCAACCACTGCTGAACCACCTCACGAACGGGTCAATACACGTTTGGATATTGAAAAAGATGATATACAAGCTGTTTTGCCGATCTCAAAGAACTGGGAGGTACTGAATACAGCCGTATTGACTGGACGTCAAGTGTCACAGTCAATGAAGGTACTCATAGTCTCTCAAGCGGGACAAGTGGCTGACGTCACATTACAGAGCTCCTGTCATTCGGAAGACGAGAGCGTTCTCAAGGTGTCATCGTCGTGCAGCTCCGTTTATGTAGATGGCTCCGAGATTCGTGGCTCATCGAACGCGTCCGTGATAGTGAAATACGGAACGTACACCGGCCTGGCAAGATTCACTGTTTGGATGCCTGAGAATCCTCTAGAAATTGATGTGGATGACAACAGATTATCCCAAATTAAAGGTTGGAAGGTGTCTGACGATGCAAGTTCAAAAGACAGACAGCGACGATCATTGTCGGCTCGAGGCTGGGGCGGATCTAGATCTGACGGCACTAACTCTTTGACGGATCAACGCTCTTGTCGATTGAGATATCAGCAGAGCAACGTAGAGGTGTACGCTCGTTTTCTTGCAAAAGATCACGACTCTGGAAGGGTGTCGTATTTCGTGTCTCGTCGAACTTGGCTTAAAGTTACCGAGCTCGTGATGTCGTTGATGCGTGTCGCCGACTTGCGCATCGCAACATTAAGAGGGAAACTTCTTCAGGGCCGGAGTACCGGCAGAACAGAGGTTCAAGTCTTATCCCCTATAACTGGTCGTGTTATCGGCGTCCGTGAAGTCCGTGTTGGTAACGACAAAGTATCGATCTCAAGGTTACTTGTTCGTGTGATATCTGGCCTTCAACTTAACATCTCTCCGGACTCGGCTATAGAAAATGGATATGTTTCCGAAACGACAGTCACGAGACGATTAACAGCACAATATCAGGAAGGTCTATTGGACATTGATATCGAGTATTCGGACGGAAGTCACACTCCGCTTCGCGATGTGGCCGTATCTGACTACTTTTTACTAGTCGAGAGCCTCGATCCCGAAGTTGTGGCTTTTGCGCCAATGTTGGCTTCCAAGCACCCACGTGTGATTGCTGTTGGTGAAGGCAGCGGTGAACTTCTTCGTGTGACTTTGTTAACGCCTGAACAATGTCGTTCCGGGCCAATACGACGCGTTGCGCTTCCCGGGAAAGGTGACGCAAAGACTCCGGCTAACATTAAAAACATTCCTGGGGCATTGGCCACGGCGGCTGCGAGCGTAGATGTCGACTTCAGCGGGGGAGATGCACCACAGAGACCGGATACGCCGCAGAACGATGATATTATGGCCAGAGGAGGACTTCGAAGTGGATTGTCGGACCTTAGCGACGTGTTGAAGGGTATATCGCCGTTCAAAGACGAAAACAACCATGAACCGACAGTGCAGGCGCAACATTACAATTCGATCTTCCGAAGCAACCCCGCGCAGCACCCCCGACAACATCAATCAACCGTCATGACGCCGGTTGAAATAGGAATGTACGTTCTCCTTGGTGCTTTCTGCTTCGCGATAGTAGTATTCGTCGTATCATGTGTGGTGTACGCTTCTCGTCTGAAGCCGGCCGGACCAGAAGGAGGCACGGGTATGTGCCGCAAGATACCGGCACTAGCTGAAGGGGGGCGGTTGCAGGTCGCAGGACTAGTGGGAACTACGCTCGGACTGGCCAGGGACAAGCCCACAAGGGAATCCACAACAAACGCTCATGATTGGGTTTGGCTAGGCAGGGCAACGATGGAGAGAAGCGGCAATCGTCATCAAGCCAATCGCAATTCCACACAAGTTCCGGACAACGGCAATCAAGAGATCAGGATCACGGCTAATCCTCTAAACTACAACTATGTTGATCCCGATGACGCGATACGAGATAACGGAAACGTTATGGTCACAAGTTTCGACAATCCAAACTCAATTGAGTTGCCATCGCAAAACGACACGAGAGTTATCGATTCCACGACTTACTGTAAGAAACCAGCTCGGCACACCCGTCAATCGTCTGGAGATGGACATTGGCAGTCGGAGCCCAACAATCCGGACGACTACCGACCGCCGGTGCCGCCACACCGACACTCCTCTACCATTCAACAGTCGCAGTTGCCCGTACCCCCACGAAATCCGAACAAAATTTTGCCCGAAGCGGTTATGCCACCAGCTAGACGAAGCCACTCAAGAAATCACAAAAAATACGAACGCGAACACGAACCCACGAAGAGATCCCCGGAAAACGCCCGCCGCTCAGACCGAATTATTGATCTAAACAAAACGGACGCACCATACGTAACGAGCAGAGATATAAGCCAGTATAAACGTGATTCGGATCCGATGCAAAACTTAAATGACTTTGATGATAACCCGGTTGAGAAACTCACGAACAAGGACGATTGCGCTCGCCTGTTTGAATTTGACAACGACGCTCCTCTGATCATGGAGAACAAGGACTACAGAGAGATAGTGGGACTGAACAGAGGTACCGACGATTCCCGACGAACATTCGCGAAGCCAGAAAGCCCTGATTACATGCACGACTCTGGAATAGGCCTACCGGACATGCAGATTCGTTATAAGAACAAAGCCAAGGAACCCAAAGGAGATTTTGTCGAGTCTGCAAACCAAAACAAACCAAGCAGCTCTCCCGAAGTGAAACGCGCCACTATAGTAGGCAACCCGATGTTCTCACGCGAGGAGCGCCGCGACCCCCCCGCGGAGGCCCTGGGACTCGACGACCTCCACATGGATTACGATCAGATCATGCACTATTTCCACAACCTCAAGGAATCGAACGCCTGA
Protein
MERCRLDVSAHLVFRELPRDSPVLRVLFHTGGEGARRAASRPRRVCITLHASLGSKSLTATCGPEGEEGACLAELTVPAAWWPADGKGKRPPRTVRLAYSATEAGNGESGEDTCGRVSVQPAWPLGSVILAGARAGYREARAGDAALLLPRAPLYPHSRIHLPFVVRRDSSHNIGHVAIRMKVKSGLRLLSVTAANSRWAVTSEVKMRSATVTATRLEQPLRDDDDIDFDEEETIGAGGPGWEEVLTWLVEVGPEGEGDAEGSEGESGRGTARLSWSAKVSPIISSSSTTAEPPHERVNTRLDIEKDDIQAVLPISKNWEVLNTAVLTGRQVSQSMKVLIVSQAGQVADVTLQSSCHSEDESVLKVSSSCSSVYVDGSEIRGSSNASVIVKYGTYTGLARFTVWMPENPLEIDVDDNRLSQIKGWKVSDDASSKDRQRRSLSARGWGGSRSDGTNSLTDQRSCRLRYQQSNVEVYARFLAKDHDSGRVSYFVSRRTWLKVTELVMSLMRVADLRIATLRGKLLQGRSTGRTEVQVLSPITGRVIGVREVRVGNDKVSISRLLVRVISGLQLNISPDSAIENGYVSETTVTRRLTAQYQEGLLDIDIEYSDGSHTPLRDVAVSDYFLLVESLDPEVVAFAPMLASKHPRVIAVGEGSGELLRVTLLTPEQCRSGPIRRVALPGKGDAKTPANIKNIPGALATAAASVDVDFSGGDAPQRPDTPQNDDIMARGGLRSGLSDLSDVLKGISPFKDENNHEPTVQAQHYNSIFRSNPAQHPRQHQSTVMTPVEIGMYVLLGAFCFAIVVFVVSCVVYASRLKPAGPEGGTGMCRKIPALAEGGRLQVAGLVGTTLGLARDKPTRESTTNAHDWVWLGRATMERSGNRHQANRNSTQVPDNGNQEIRITANPLNYNYVDPDDAIRDNGNVMVTSFDNPNSIELPSQNDTRVIDSTTYCKKPARHTRQSSGDGHWQSEPNNPDDYRPPVPPHRHSSTIQQSQLPVPPRNPNKILPEAVMPPARRSHSRNHKKYEREHEPTKRSPENARRSDRIIDLNKTDAPYVTSRDISQYKRDSDPMQNLNDFDDNPVEKLTNKDDCARLFEFDNDAPLIMENKDYREIVGLNRGTDDSRRTFAKPESPDYMHDSGIGLPDMQIRYKNKAKEPKGDFVESANQNKPSSSPEVKRATIVGNPMFSREERRDPPAEALGLDDLHMDYDQIMHYFHNLKESNA
Summary
Uniprot
H9JBP0
A0A2H1WH03
A0A194QEA0
A0A2W1BZ92
A0A194QS66
A0A1Y1MJ17
+ More
A0A1Y1MQT4 A0A1Y1MMI2 A0A1Y1MNY2 A0A1Y1MKW9 A0A1Y1MIV7 A0A182GW34 A0A1B6D587 A0A1S4F8W6 Q17BQ2 A0A1Q3FPX6 A0A1Q3FPT9 A0A1Q3FPQ3 A0A1Q3FPY9 A0A1Q3FQ78 A0A0P6K0P2 A0A1J1I421 A0A1B6FRB1 W5J6D9 A0A1B6G8D2 B0W1X2 Q7PMN0 A0A1S4J250 A0A1S4GMD1 A0A182F8T7 A0A182K5L3 A0A084VYR8 A0A151I0C4 A0A182RKU2 A0A182YEM9 A0A195CV35 A0A232FCE0 A0A0A1XPB0 A0A151X924 Q29GG0 U4UA03 B3NXP6 A0A3B0K0X4 A0A3B0JZ35 A0A1W4VS41 A0A1A9VC73 X2JDW6 A0A1B0A9W6 A0A1B0FGG5 A0A1W4VTF4 X2JEB0 Q9W427 B4R5E0 A0A1W4VF40 B4PYP9 B4I0D9 A0A0R1E8Z9 A0A088AA12 A0A1A9WFW8 A0A0K8VK21 B4NPD6 W8BNH8 B3MQ58 A0A0M3QYY4 A0A0N0BKF3 B4MAJ3 A0A1I8Q7X7 B4L672 A0A0K8VPK6 A0A0L0CG47 A0A1B6K692 B4JM03 B4HB95 A0A182HWY4 A0A023F0K4 A0A1A9YPK0 A0A1B0BJV1 A0A182WYV9
A0A1Y1MQT4 A0A1Y1MMI2 A0A1Y1MNY2 A0A1Y1MKW9 A0A1Y1MIV7 A0A182GW34 A0A1B6D587 A0A1S4F8W6 Q17BQ2 A0A1Q3FPX6 A0A1Q3FPT9 A0A1Q3FPQ3 A0A1Q3FPY9 A0A1Q3FQ78 A0A0P6K0P2 A0A1J1I421 A0A1B6FRB1 W5J6D9 A0A1B6G8D2 B0W1X2 Q7PMN0 A0A1S4J250 A0A1S4GMD1 A0A182F8T7 A0A182K5L3 A0A084VYR8 A0A151I0C4 A0A182RKU2 A0A182YEM9 A0A195CV35 A0A232FCE0 A0A0A1XPB0 A0A151X924 Q29GG0 U4UA03 B3NXP6 A0A3B0K0X4 A0A3B0JZ35 A0A1W4VS41 A0A1A9VC73 X2JDW6 A0A1B0A9W6 A0A1B0FGG5 A0A1W4VTF4 X2JEB0 Q9W427 B4R5E0 A0A1W4VF40 B4PYP9 B4I0D9 A0A0R1E8Z9 A0A088AA12 A0A1A9WFW8 A0A0K8VK21 B4NPD6 W8BNH8 B3MQ58 A0A0M3QYY4 A0A0N0BKF3 B4MAJ3 A0A1I8Q7X7 B4L672 A0A0K8VPK6 A0A0L0CG47 A0A1B6K692 B4JM03 B4HB95 A0A182HWY4 A0A023F0K4 A0A1A9YPK0 A0A1B0BJV1 A0A182WYV9
Pubmed
EMBL
BABH01024436
BABH01024437
BABH01024438
ODYU01008616
SOQ52351.1
KQ459302
+ More
KPJ01791.1 KZ149900 PZC78567.1 KQ461175 KPJ07825.1 GEZM01030278 JAV85653.1 GEZM01030274 JAV85657.1 GEZM01030276 JAV85655.1 GEZM01030275 JAV85656.1 GEZM01030273 JAV85658.1 GEZM01030277 JAV85654.1 JXUM01092324 JXUM01092325 KQ563979 KXJ72988.1 GEDC01016444 JAS20854.1 CH477318 EAT43715.1 GFDL01005461 JAV29584.1 GFDL01005426 JAV29619.1 GFDL01005454 JAV29591.1 GFDL01005502 JAV29543.1 GFDL01005462 JAV29583.1 GDUN01001047 JAN94872.1 CVRI01000040 CRK95087.1 GECZ01017216 JAS52553.1 ADMH02002005 ETN60007.1 GECZ01011051 JAS58718.1 DS231824 EDS27408.1 AAAB01008977 EAA13567.6 ATLV01018422 KE525233 KFB43112.1 KQ976639 KYM78825.1 KQ977279 KYN04024.1 NNAY01000452 OXU28292.1 GBXI01001899 JAD12393.1 KQ982398 KYQ56871.1 CH379064 EAL32149.3 KB631865 ERL86780.1 CH954180 EDV47347.2 OUUW01000011 SPP86332.1 SPP86333.1 AE014298 AHN59403.1 CCAG010007471 AHN59404.1 AAF46133.2 AFH07261.1 CM000366 EDX17236.1 CM000162 EDX00985.2 CH480819 EDW52970.1 KRK05864.1 GDHF01013389 JAI38925.1 CH964291 EDW86376.2 GAMC01003910 JAC02646.1 CH902621 EDV44484.2 CP012528 ALC48483.1 KQ435700 KOX80512.1 CH940655 EDW66252.2 CH933812 EDW05868.2 GDHF01011493 JAI40821.1 JRES01000520 KNC30454.1 GECU01000736 JAT06971.1 CH916371 EDV91764.1 CH479248 EDW37907.1 APCN01008384 GBBI01004218 JAC14494.1 JXJN01015633 JXJN01015634 JXJN01015635
KPJ01791.1 KZ149900 PZC78567.1 KQ461175 KPJ07825.1 GEZM01030278 JAV85653.1 GEZM01030274 JAV85657.1 GEZM01030276 JAV85655.1 GEZM01030275 JAV85656.1 GEZM01030273 JAV85658.1 GEZM01030277 JAV85654.1 JXUM01092324 JXUM01092325 KQ563979 KXJ72988.1 GEDC01016444 JAS20854.1 CH477318 EAT43715.1 GFDL01005461 JAV29584.1 GFDL01005426 JAV29619.1 GFDL01005454 JAV29591.1 GFDL01005502 JAV29543.1 GFDL01005462 JAV29583.1 GDUN01001047 JAN94872.1 CVRI01000040 CRK95087.1 GECZ01017216 JAS52553.1 ADMH02002005 ETN60007.1 GECZ01011051 JAS58718.1 DS231824 EDS27408.1 AAAB01008977 EAA13567.6 ATLV01018422 KE525233 KFB43112.1 KQ976639 KYM78825.1 KQ977279 KYN04024.1 NNAY01000452 OXU28292.1 GBXI01001899 JAD12393.1 KQ982398 KYQ56871.1 CH379064 EAL32149.3 KB631865 ERL86780.1 CH954180 EDV47347.2 OUUW01000011 SPP86332.1 SPP86333.1 AE014298 AHN59403.1 CCAG010007471 AHN59404.1 AAF46133.2 AFH07261.1 CM000366 EDX17236.1 CM000162 EDX00985.2 CH480819 EDW52970.1 KRK05864.1 GDHF01013389 JAI38925.1 CH964291 EDW86376.2 GAMC01003910 JAC02646.1 CH902621 EDV44484.2 CP012528 ALC48483.1 KQ435700 KOX80512.1 CH940655 EDW66252.2 CH933812 EDW05868.2 GDHF01011493 JAI40821.1 JRES01000520 KNC30454.1 GECU01000736 JAT06971.1 CH916371 EDV91764.1 CH479248 EDW37907.1 APCN01008384 GBBI01004218 JAC14494.1 JXJN01015633 JXJN01015634 JXJN01015635
Proteomes
UP000005204
UP000053268
UP000053240
UP000069940
UP000249989
UP000008820
+ More
UP000183832 UP000000673 UP000002320 UP000007062 UP000069272 UP000075881 UP000030765 UP000078540 UP000075900 UP000076408 UP000078542 UP000215335 UP000075809 UP000001819 UP000030742 UP000008711 UP000268350 UP000192221 UP000078200 UP000000803 UP000092445 UP000092444 UP000000304 UP000002282 UP000001292 UP000005203 UP000091820 UP000007798 UP000007801 UP000092553 UP000053105 UP000008792 UP000095300 UP000009192 UP000037069 UP000001070 UP000008744 UP000075840 UP000092443 UP000092460 UP000076407
UP000183832 UP000000673 UP000002320 UP000007062 UP000069272 UP000075881 UP000030765 UP000078540 UP000075900 UP000076408 UP000078542 UP000215335 UP000075809 UP000001819 UP000030742 UP000008711 UP000268350 UP000192221 UP000078200 UP000000803 UP000092445 UP000092444 UP000000304 UP000002282 UP000001292 UP000005203 UP000091820 UP000007798 UP000007801 UP000092553 UP000053105 UP000008792 UP000095300 UP000009192 UP000037069 UP000001070 UP000008744 UP000075840 UP000092443 UP000092460 UP000076407
Interpro
SUPFAM
SSF48619
SSF48619
Gene 3D
ProteinModelPortal
H9JBP0
A0A2H1WH03
A0A194QEA0
A0A2W1BZ92
A0A194QS66
A0A1Y1MJ17
+ More
A0A1Y1MQT4 A0A1Y1MMI2 A0A1Y1MNY2 A0A1Y1MKW9 A0A1Y1MIV7 A0A182GW34 A0A1B6D587 A0A1S4F8W6 Q17BQ2 A0A1Q3FPX6 A0A1Q3FPT9 A0A1Q3FPQ3 A0A1Q3FPY9 A0A1Q3FQ78 A0A0P6K0P2 A0A1J1I421 A0A1B6FRB1 W5J6D9 A0A1B6G8D2 B0W1X2 Q7PMN0 A0A1S4J250 A0A1S4GMD1 A0A182F8T7 A0A182K5L3 A0A084VYR8 A0A151I0C4 A0A182RKU2 A0A182YEM9 A0A195CV35 A0A232FCE0 A0A0A1XPB0 A0A151X924 Q29GG0 U4UA03 B3NXP6 A0A3B0K0X4 A0A3B0JZ35 A0A1W4VS41 A0A1A9VC73 X2JDW6 A0A1B0A9W6 A0A1B0FGG5 A0A1W4VTF4 X2JEB0 Q9W427 B4R5E0 A0A1W4VF40 B4PYP9 B4I0D9 A0A0R1E8Z9 A0A088AA12 A0A1A9WFW8 A0A0K8VK21 B4NPD6 W8BNH8 B3MQ58 A0A0M3QYY4 A0A0N0BKF3 B4MAJ3 A0A1I8Q7X7 B4L672 A0A0K8VPK6 A0A0L0CG47 A0A1B6K692 B4JM03 B4HB95 A0A182HWY4 A0A023F0K4 A0A1A9YPK0 A0A1B0BJV1 A0A182WYV9
A0A1Y1MQT4 A0A1Y1MMI2 A0A1Y1MNY2 A0A1Y1MKW9 A0A1Y1MIV7 A0A182GW34 A0A1B6D587 A0A1S4F8W6 Q17BQ2 A0A1Q3FPX6 A0A1Q3FPT9 A0A1Q3FPQ3 A0A1Q3FPY9 A0A1Q3FQ78 A0A0P6K0P2 A0A1J1I421 A0A1B6FRB1 W5J6D9 A0A1B6G8D2 B0W1X2 Q7PMN0 A0A1S4J250 A0A1S4GMD1 A0A182F8T7 A0A182K5L3 A0A084VYR8 A0A151I0C4 A0A182RKU2 A0A182YEM9 A0A195CV35 A0A232FCE0 A0A0A1XPB0 A0A151X924 Q29GG0 U4UA03 B3NXP6 A0A3B0K0X4 A0A3B0JZ35 A0A1W4VS41 A0A1A9VC73 X2JDW6 A0A1B0A9W6 A0A1B0FGG5 A0A1W4VTF4 X2JEB0 Q9W427 B4R5E0 A0A1W4VF40 B4PYP9 B4I0D9 A0A0R1E8Z9 A0A088AA12 A0A1A9WFW8 A0A0K8VK21 B4NPD6 W8BNH8 B3MQ58 A0A0M3QYY4 A0A0N0BKF3 B4MAJ3 A0A1I8Q7X7 B4L672 A0A0K8VPK6 A0A0L0CG47 A0A1B6K692 B4JM03 B4HB95 A0A182HWY4 A0A023F0K4 A0A1A9YPK0 A0A1B0BJV1 A0A182WYV9
Ontologies
PANTHER
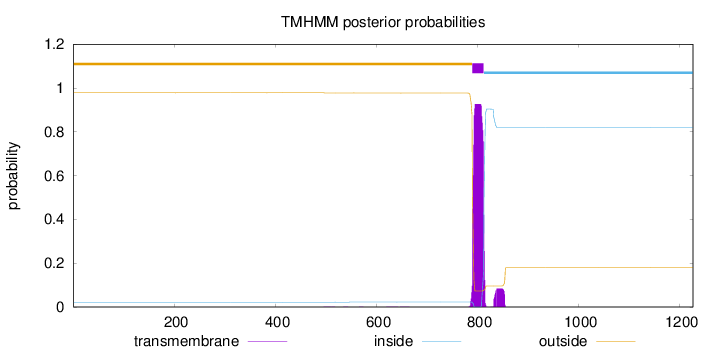
Topology
Length:
1226
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.66667
Exp number, first 60 AAs:
0.00051
Total prob of N-in:
0.02197
outside
1 - 789
TMhelix
790 - 812
inside
813 - 1226
Population Genetic Test Statistics
Pi
229.047313
Theta
184.716572
Tajima's D
0.191608
CLR
0.428229
CSRT
0.424128793560322
Interpretation
Uncertain
