Pre Gene Modal
BGIBMGA006920
Annotation
Leucine-rich_repeats_and_immunoglobulin-like_domains_protein_3_[Papilio_machaon]
Full name
Leucine-rich repeats and immunoglobulin-like domains protein 3
Location in the cell
Nuclear Reliability : 1.689 PlasmaMembrane Reliability : 1.164
Sequence
CDS
ATGGGAGACTCTCGATTGTTATTGTTTATAACAATGAGCTTGTTCCTTAGTTCATCGACTCTGGGGGAAATTTACTTTTGCCCATTAGAATGCGCCTGTCTGCATTTATATGTCGATTGTTCAAAGAAGAATTTGGTGTCTGTGCCAAAAATTCCTGAGTGGGCGGAGCAATTGGACCTCAGTCATAATAAACTAAATAATGTCACGAATGAATTGAATACGCTATCGAATTTAAAAGTATTAAAACTCGACAAGAATAATATGAACAATATACCGAACCTCCAGTTGGACGAAATACTAGAACTAAACCTCAATAGAAATAACATTGTATCAATAGACACGAATCATTTTCCGGAAAATAGCTCTTTGAAAATATTAAATCTCAACAGTAATCACATAATGACGATACATGAAGGAGCTATTGATAATTTATCCAACTTGGAAGTATTGAAATTAAACAGAAATGAGATTAAATCTCTCCCAAATCATTTATTCAAGTATCAAACAAATTTGAGAATTCTAGAACTGAATCACAATAAATTGCATGTTATCAATGCACTGCTTTTTCAAGGATTATCAAATTTGACTACTTTGAAACTTAGATTTAACAACATAGATAAAATAATGGATGGTGCCTTTTTCGGATTTAAATCAATGAACCTATTGCAATTGGATCATAATAAAATCAGAGGTATATCAAAAGGGTGGATGTATGGCCTTGAATCTTTGACAACATTATCTATGTCCAACAACTTGATATCTAATATTGATCTAGGAAGCTGGGAACTGTGTGCAAATTTAGAGGTTCTTGATTTATCACACAATCATTTGATAGAAATAGAGGGGCGTACCTTTCAACACCTAATAAACCTTGAAACATTAAATCTTAGTAACAATAATATATCATCTATTTCACAAGAGTCCTTCAGTTATATGGCTCATCTTAAAGCACTTTATTTGAATAACAACAAAATTTCTTGGACTGTAGAAGACATGTCTGGGCCTTTCTCAAAATTACAAAATTTACACTTCTTTAACTTAGCAAGTAATCATATAAAGTCTATAAACTCCAGGGCATTTGATGGTCTTTCCAATCTATTGGAGTTAAATTTGGCTGGAAATAACATAACATCTATTCAGCAGCATGCATTTAATTCTATGACAAACCTGAAAAAACTACACATGAACTCGTCATCGCTACTCTGTGACTGTAATTTGTTATGGATTGTTAAATGGATAAAAGAAAAAGCTGAGCAAAAATTTATCACTGCTACTTGTGCATACCCCTCTGAAGTCAGAGGTGTGTCAATTACAAAACTGAAAGAGGAAAACTGTGGAGATTCGCCAAAACCAAAAATTGTAGAGCATCCTAAAACACAGCTTGCAATTAAAGGGCGTTCTGCTAGTCTAAGTGGTCAACCTCGTGCAACATCTACCCTAATTATACCTAACATCTCACAAGGAGATTCTGGTAAATACCAATGTGTTGTAACAAATAATTTTGGAACTACATATTCTTCAAAAGCAAAAATGAATATTGTCACATTCCCTAGGTTTATAAAAACACCAACAAATGTGACTGTTAAAACTGGTGAAACTGTCACACTGAACTGTGCAGCTACTGGAGACCCTACTCCAGAGATATCATGGAAAAAAGATGGGGGGAATGACTTCCCTGCAGCTAGAGAAAGAAGAATGAATGTTATGCCGGATGATCATTTATTTTTTATTGTTAATGTAAAGACAACAGATATTGGTGTTTACAGTTGCGCTGCAAAAAATCCTGCCGGCACTATTGTGGCTAATGCTACATTGACAGTATTACAAGAACCTTCATTTGCTAAGGTAATGGAAAACAAAGAAGTCATCAGTGGAGACTCAGTTGTTTTACAATGTATGATTTCGGGATCACCTAAGCCAGTTTTGAAATGGCTAAAAGATGATACTCCAGTTGTCCCTGGCGAAAGACATTTCTTAGTTAATGATGACCAACTATTGATCATAACAGGAGTACAATCTGACGATGCTGGTAAATATGATTGTGAGATCACAAATGAACTGGGAACTAAGAAACATATGACTCAGCTACAAGTATTGCCACCTGTAGCAATTATGGTTAATGAGGAAGATATGACTGGCATCATAATTATAACTGTAGTGTGTTGTGCTGTTGGAACTTCAATAATATGGGTAGTAATTATATATCATACCCGTAGGCGTATGGCAACTGCAGTGCGTATTCCTACAGAAAGTTTGAAAAGAACACATATAGTGCACAGTGACAGTGATTCTCCACAAATGTTTTCAGACAATGTTTCAGAACATTCATCATGTAAGGATAGTGGGACTGGTGATTCTGCTAAACAAACGAGCTTTGATGGTGTTCCTATGGAAAGGATTGAACAAGTACACTTTGAGCCTCCTGCCACCAACCGGAGCTATTCTCCTGTGCCTGAGGCTCATAAACTTCTTCCATCTTCCTTTAAACCATCAATTCAATCCGGATATCCAGGACAGGCAGCTTACGGTGGAATGCCACCAGGCCAATTAGAAATTGGACATGGACCGTACCCTTCCATTGGAGTTGGGGGTACTATTACGCCCCAAGTACAGCAGTGGTTCCGAGCTGTAGATAAAGATCAATCTGGATTCATTACTTCGACAGAACTAAGATCAGCGTTGGTGAACGCTCAAGGTCAAACTTTTTCCGAAACTGCTTGCAATCTGATGATTGGTATGTTCGACAAAGACCGCAACGGGCAGATTAATCTCGATGAATTTGATAAGCTTTATACTTACGTGAATCAATGGTTAGCTGTATTCAAAACCTACGATACGGATCAATCTGGTTTCATAGATGAAGGCGAACTTTCTAAAGCTCTGACACAAATGGGATTCCGGTTCTCACCAGAGTTTATAAATATTTTAACGAAACGGAGTAACACAAATGATGGAAAAATTTCCGTGGACCAGTTTATTGTGATGTGCGTTCAGATACAACGTTTCACTGAAGCATTTCGAGTACGCGATACCGAACAAAATGGGACGGTTACCATAGCCTTTGAAGACTTCCTTACTATTGCCCTGAGCTGTTCTATTTGA
Protein
MGDSRLLLFITMSLFLSSSTLGEIYFCPLECACLHLYVDCSKKNLVSVPKIPEWAEQLDLSHNKLNNVTNELNTLSNLKVLKLDKNNMNNIPNLQLDEILELNLNRNNIVSIDTNHFPENSSLKILNLNSNHIMTIHEGAIDNLSNLEVLKLNRNEIKSLPNHLFKYQTNLRILELNHNKLHVINALLFQGLSNLTTLKLRFNNIDKIMDGAFFGFKSMNLLQLDHNKIRGISKGWMYGLESLTTLSMSNNLISNIDLGSWELCANLEVLDLSHNHLIEIEGRTFQHLINLETLNLSNNNISSISQESFSYMAHLKALYLNNNKISWTVEDMSGPFSKLQNLHFFNLASNHIKSINSRAFDGLSNLLELNLAGNNITSIQQHAFNSMTNLKKLHMNSSSLLCDCNLLWIVKWIKEKAEQKFITATCAYPSEVRGVSITKLKEENCGDSPKPKIVEHPKTQLAIKGRSASLSGQPRATSTLIIPNISQGDSGKYQCVVTNNFGTTYSSKAKMNIVTFPRFIKTPTNVTVKTGETVTLNCAATGDPTPEISWKKDGGNDFPAARERRMNVMPDDHLFFIVNVKTTDIGVYSCAAKNPAGTIVANATLTVLQEPSFAKVMENKEVISGDSVVLQCMISGSPKPVLKWLKDDTPVVPGERHFLVNDDQLLIITGVQSDDAGKYDCEITNELGTKKHMTQLQVLPPVAIMVNEEDMTGIIIITVVCCAVGTSIIWVVIIYHTRRRMATAVRIPTESLKRTHIVHSDSDSPQMFSDNVSEHSSCKDSGTGDSAKQTSFDGVPMERIEQVHFEPPATNRSYSPVPEAHKLLPSSFKPSIQSGYPGQAAYGGMPPGQLEIGHGPYPSIGVGGTITPQVQQWFRAVDKDQSGFITSTELRSALVNAQGQTFSETACNLMIGMFDKDRNGQINLDEFDKLYTYVNQWLAVFKTYDTDQSGFIDEGELSKALTQMGFRFSPEFINILTKRSNTNDGKISVDQFIVMCVQIQRFTEAFRVRDTEQNGTVTIAFEDFLTIALSCSI
Summary
Description
May play a role in craniofacial and inner ear morphogenesis during embryonic development. May act within the otic vesicle epithelium to control formation of the lateral semicircular canal in the inner ear, possibly by restricting the expression of NTN1 (By similarity).
Subunit
Interacts with EGFR, ERBB2 and ERBB4 (in vitro).
Keywords
Alternative splicing
Cell membrane
Complete proteome
Cytoplasmic vesicle
Developmental protein
Disulfide bond
Glycoprotein
Immunoglobulin domain
Leucine-rich repeat
Membrane
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Feature
chain Leucine-rich repeats and immunoglobulin-like domains protein 3
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A194RBK8
A0A194QEA5
A0A0L7LRL8
A0A2H1W9I2
A0A2A4JQA6
A0A2W1BU19
+ More
A0A194RCI6 A0A2J7PXZ3 A0A2J7PY00 A0A2P8ZLE5 E2BGQ9 A0A0N8A6G9 A0A0P5HNG9 A0A026VY36 D6W6K8 A0A0J7NT49 A0A088AUX2 A0A146L7E5 A0A0P6AM67 A0A164W9T3 A0A0P6GUV0 A0A2A3E803 E1ZVE8 V4BQX8 A0A1B6GRF4 A0A1B6E3N8 R7V016 K7IXQ9 A0A151WKY9 A0A0T6BC00 A0A195BGE0 A0A195DQZ1 A0A195FD24 I3M049 F4X8H7 H0VJB6 A0A151IIY4 A0A158NBN2 F6W7K8 A0A2K6F4G2 A0A3Q7WZW6 A0A2Y9GYK9 A0A3Q7V3Q9 G3SKZ9 G3U7I2 G3RJB5 A0A0P5RBF1 A0A2U3X4L7 D2H328 H2NHW0 Q6UXM1 H2Q6D1 A0A2Y9KYJ8 G1S5Q1 A0A3Q7PYL0 E1BN69 A0A2Y9FNH4 M3XSK0 F1SKD7 A0A383ZXC5 G1T9T3 F6YDB3 A0A232ES86 A0A1S2ZBA6 A0A2J8KVQ9 W5NIH5 W5NIH3 A0A1S3FF81 A0A1L8GYN2 A0A1L8GUD1 A0A287AI53 A0A3Q3KMT6 H3CN76 A0A1S3SHC9 A0A3B4CUF8 A0A3B3TED3 A0A3Q3KMV7 A0A1S3N0F3 A0A3Q1F4B6 A0A147BU85 A0A131XVU3 W5P3P1 A0A131XF45 W5LKM1 A0A1B6MI28 A0A1B6KHJ9 L7MIH9 I3JUG3
A0A194RCI6 A0A2J7PXZ3 A0A2J7PY00 A0A2P8ZLE5 E2BGQ9 A0A0N8A6G9 A0A0P5HNG9 A0A026VY36 D6W6K8 A0A0J7NT49 A0A088AUX2 A0A146L7E5 A0A0P6AM67 A0A164W9T3 A0A0P6GUV0 A0A2A3E803 E1ZVE8 V4BQX8 A0A1B6GRF4 A0A1B6E3N8 R7V016 K7IXQ9 A0A151WKY9 A0A0T6BC00 A0A195BGE0 A0A195DQZ1 A0A195FD24 I3M049 F4X8H7 H0VJB6 A0A151IIY4 A0A158NBN2 F6W7K8 A0A2K6F4G2 A0A3Q7WZW6 A0A2Y9GYK9 A0A3Q7V3Q9 G3SKZ9 G3U7I2 G3RJB5 A0A0P5RBF1 A0A2U3X4L7 D2H328 H2NHW0 Q6UXM1 H2Q6D1 A0A2Y9KYJ8 G1S5Q1 A0A3Q7PYL0 E1BN69 A0A2Y9FNH4 M3XSK0 F1SKD7 A0A383ZXC5 G1T9T3 F6YDB3 A0A232ES86 A0A1S2ZBA6 A0A2J8KVQ9 W5NIH5 W5NIH3 A0A1S3FF81 A0A1L8GYN2 A0A1L8GUD1 A0A287AI53 A0A3Q3KMT6 H3CN76 A0A1S3SHC9 A0A3B4CUF8 A0A3B3TED3 A0A3Q3KMV7 A0A1S3N0F3 A0A3Q1F4B6 A0A147BU85 A0A131XVU3 W5P3P1 A0A131XF45 W5LKM1 A0A1B6MI28 A0A1B6KHJ9 L7MIH9 I3JUG3
Pubmed
26354079
26227816
28756777
29403074
20798317
24508170
+ More
30249741 18362917 19820115 26823975 23254933 20075255 21719571 21993624 21347285 19892987 22398555 20010809 15203213 12975309 14702039 16136131 19393038 20431018 28648823 27762356 15496914 29240929 29652888 20809919 28049606 25329095 25576852 25186727
30249741 18362917 19820115 26823975 23254933 20075255 21719571 21993624 21347285 19892987 22398555 20010809 15203213 12975309 14702039 16136131 19393038 20431018 28648823 27762356 15496914 29240929 29652888 20809919 28049606 25329095 25576852 25186727
EMBL
KQ460398
KPJ14997.1
KQ459302
KPJ01796.1
JTDY01000239
KOB78115.1
+ More
ODYU01006887 SOQ49134.1 NWSH01000905 PCG73590.1 KZ149900 PZC78572.1 KPJ14995.1 NEVH01020852 PNF21218.1 PNF21219.1 PYGN01000023 PSN57328.1 GL448204 EFN85129.1 GDIP01177785 JAJ45617.1 GDIQ01230832 JAK20893.1 KK107638 QOIP01000004 EZA48386.1 RLU23208.1 KQ971307 EFA10831.1 LBMM01001902 KMQ95575.1 GDHC01015537 JAQ03092.1 GDIP01027507 JAM76208.1 LRGB01001274 KZS13085.1 GDIQ01028504 JAN66233.1 KZ288356 PBC27181.1 GL434492 EFN74858.1 KB202283 ESO91319.1 GECZ01004789 JAS64980.1 GEDC01004745 JAS32553.1 AMQN01005574 AMQN01005575 AMQN01005576 KB296320 ELU11894.1 KQ982981 KYQ48549.1 LJIG01002086 KRT84854.1 KQ976500 KYM83242.1 KQ980581 KYN15330.1 KQ981673 KYN38308.1 AGTP01085485 AGTP01085486 AGTP01085487 GL888932 EGI57246.1 AAKN02014756 KQ977445 KYN02610.1 ADTU01011228 CABD030083438 CABD030083439 GDIQ01122668 JAL29058.1 ACTA01002073 ACTA01010073 GL192457 EFB28898.1 ABGA01160822 ABGA01160823 ABGA01160824 ABGA01160825 ABGA01160826 ABGA01160827 NDHI03003457 PNJ44445.1 AY505340 AY358288 AY358295 AK074921 AACZ04012982 ADFV01002192 ADFV01002193 AEYP01080416 AEYP01080417 AEMK02000033 AAMC01103647 AAMC01103648 NNAY01002474 OXU21223.1 NBAG03000333 PNI39111.1 AHAT01037175 CM004470 OCT88950.1 CM004471 OCT87435.1 GEGO01001101 JAR94303.1 GEFM01004383 JAP71413.1 AMGL01081444 AMGL01081445 GEFH01003609 JAP64972.1 GEBQ01004448 JAT35529.1 GEBQ01029343 JAT10634.1 GACK01002040 JAA62994.1 AERX01012898 AERX01012899
ODYU01006887 SOQ49134.1 NWSH01000905 PCG73590.1 KZ149900 PZC78572.1 KPJ14995.1 NEVH01020852 PNF21218.1 PNF21219.1 PYGN01000023 PSN57328.1 GL448204 EFN85129.1 GDIP01177785 JAJ45617.1 GDIQ01230832 JAK20893.1 KK107638 QOIP01000004 EZA48386.1 RLU23208.1 KQ971307 EFA10831.1 LBMM01001902 KMQ95575.1 GDHC01015537 JAQ03092.1 GDIP01027507 JAM76208.1 LRGB01001274 KZS13085.1 GDIQ01028504 JAN66233.1 KZ288356 PBC27181.1 GL434492 EFN74858.1 KB202283 ESO91319.1 GECZ01004789 JAS64980.1 GEDC01004745 JAS32553.1 AMQN01005574 AMQN01005575 AMQN01005576 KB296320 ELU11894.1 KQ982981 KYQ48549.1 LJIG01002086 KRT84854.1 KQ976500 KYM83242.1 KQ980581 KYN15330.1 KQ981673 KYN38308.1 AGTP01085485 AGTP01085486 AGTP01085487 GL888932 EGI57246.1 AAKN02014756 KQ977445 KYN02610.1 ADTU01011228 CABD030083438 CABD030083439 GDIQ01122668 JAL29058.1 ACTA01002073 ACTA01010073 GL192457 EFB28898.1 ABGA01160822 ABGA01160823 ABGA01160824 ABGA01160825 ABGA01160826 ABGA01160827 NDHI03003457 PNJ44445.1 AY505340 AY358288 AY358295 AK074921 AACZ04012982 ADFV01002192 ADFV01002193 AEYP01080416 AEYP01080417 AEMK02000033 AAMC01103647 AAMC01103648 NNAY01002474 OXU21223.1 NBAG03000333 PNI39111.1 AHAT01037175 CM004470 OCT88950.1 CM004471 OCT87435.1 GEGO01001101 JAR94303.1 GEFM01004383 JAP71413.1 AMGL01081444 AMGL01081445 GEFH01003609 JAP64972.1 GEBQ01004448 JAT35529.1 GEBQ01029343 JAT10634.1 GACK01002040 JAA62994.1 AERX01012898 AERX01012899
Proteomes
UP000053240
UP000053268
UP000037510
UP000218220
UP000235965
UP000245037
+ More
UP000008237 UP000053097 UP000279307 UP000007266 UP000036403 UP000005203 UP000076858 UP000242457 UP000000311 UP000030746 UP000014760 UP000002358 UP000075809 UP000078540 UP000078492 UP000078541 UP000005215 UP000007755 UP000005447 UP000078542 UP000005205 UP000002281 UP000233160 UP000286642 UP000248481 UP000007646 UP000001519 UP000245340 UP000008912 UP000001595 UP000005640 UP000002277 UP000248482 UP000001073 UP000286641 UP000009136 UP000248484 UP000000715 UP000008227 UP000261681 UP000001811 UP000008143 UP000215335 UP000079721 UP000018468 UP000081671 UP000186698 UP000261640 UP000007303 UP000087266 UP000261440 UP000261540 UP000257200 UP000002356 UP000018467 UP000005207
UP000008237 UP000053097 UP000279307 UP000007266 UP000036403 UP000005203 UP000076858 UP000242457 UP000000311 UP000030746 UP000014760 UP000002358 UP000075809 UP000078540 UP000078492 UP000078541 UP000005215 UP000007755 UP000005447 UP000078542 UP000005205 UP000002281 UP000233160 UP000286642 UP000248481 UP000007646 UP000001519 UP000245340 UP000008912 UP000001595 UP000005640 UP000002277 UP000248482 UP000001073 UP000286641 UP000009136 UP000248484 UP000000715 UP000008227 UP000261681 UP000001811 UP000008143 UP000215335 UP000079721 UP000018468 UP000081671 UP000186698 UP000261640 UP000007303 UP000087266 UP000261440 UP000261540 UP000257200 UP000002356 UP000018467 UP000005207
Pfam
Interpro
IPR003599
Ig_sub
+ More
IPR000483 Cys-rich_flank_reg_C
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR032675 LRR_dom_sf
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR013098 Ig_I-set
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR003598 Ig_sub2
IPR018247 EF_Hand_1_Ca_BS
IPR000372 LRRNT
IPR036179 Ig-like_dom_sf
IPR026906 LRR_5
IPR000483 Cys-rich_flank_reg_C
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR032675 LRR_dom_sf
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR013098 Ig_I-set
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR003598 Ig_sub2
IPR018247 EF_Hand_1_Ca_BS
IPR000372 LRRNT
IPR036179 Ig-like_dom_sf
IPR026906 LRR_5
Gene 3D
ProteinModelPortal
A0A194RBK8
A0A194QEA5
A0A0L7LRL8
A0A2H1W9I2
A0A2A4JQA6
A0A2W1BU19
+ More
A0A194RCI6 A0A2J7PXZ3 A0A2J7PY00 A0A2P8ZLE5 E2BGQ9 A0A0N8A6G9 A0A0P5HNG9 A0A026VY36 D6W6K8 A0A0J7NT49 A0A088AUX2 A0A146L7E5 A0A0P6AM67 A0A164W9T3 A0A0P6GUV0 A0A2A3E803 E1ZVE8 V4BQX8 A0A1B6GRF4 A0A1B6E3N8 R7V016 K7IXQ9 A0A151WKY9 A0A0T6BC00 A0A195BGE0 A0A195DQZ1 A0A195FD24 I3M049 F4X8H7 H0VJB6 A0A151IIY4 A0A158NBN2 F6W7K8 A0A2K6F4G2 A0A3Q7WZW6 A0A2Y9GYK9 A0A3Q7V3Q9 G3SKZ9 G3U7I2 G3RJB5 A0A0P5RBF1 A0A2U3X4L7 D2H328 H2NHW0 Q6UXM1 H2Q6D1 A0A2Y9KYJ8 G1S5Q1 A0A3Q7PYL0 E1BN69 A0A2Y9FNH4 M3XSK0 F1SKD7 A0A383ZXC5 G1T9T3 F6YDB3 A0A232ES86 A0A1S2ZBA6 A0A2J8KVQ9 W5NIH5 W5NIH3 A0A1S3FF81 A0A1L8GYN2 A0A1L8GUD1 A0A287AI53 A0A3Q3KMT6 H3CN76 A0A1S3SHC9 A0A3B4CUF8 A0A3B3TED3 A0A3Q3KMV7 A0A1S3N0F3 A0A3Q1F4B6 A0A147BU85 A0A131XVU3 W5P3P1 A0A131XF45 W5LKM1 A0A1B6MI28 A0A1B6KHJ9 L7MIH9 I3JUG3
A0A194RCI6 A0A2J7PXZ3 A0A2J7PY00 A0A2P8ZLE5 E2BGQ9 A0A0N8A6G9 A0A0P5HNG9 A0A026VY36 D6W6K8 A0A0J7NT49 A0A088AUX2 A0A146L7E5 A0A0P6AM67 A0A164W9T3 A0A0P6GUV0 A0A2A3E803 E1ZVE8 V4BQX8 A0A1B6GRF4 A0A1B6E3N8 R7V016 K7IXQ9 A0A151WKY9 A0A0T6BC00 A0A195BGE0 A0A195DQZ1 A0A195FD24 I3M049 F4X8H7 H0VJB6 A0A151IIY4 A0A158NBN2 F6W7K8 A0A2K6F4G2 A0A3Q7WZW6 A0A2Y9GYK9 A0A3Q7V3Q9 G3SKZ9 G3U7I2 G3RJB5 A0A0P5RBF1 A0A2U3X4L7 D2H328 H2NHW0 Q6UXM1 H2Q6D1 A0A2Y9KYJ8 G1S5Q1 A0A3Q7PYL0 E1BN69 A0A2Y9FNH4 M3XSK0 F1SKD7 A0A383ZXC5 G1T9T3 F6YDB3 A0A232ES86 A0A1S2ZBA6 A0A2J8KVQ9 W5NIH5 W5NIH3 A0A1S3FF81 A0A1L8GYN2 A0A1L8GUD1 A0A287AI53 A0A3Q3KMT6 H3CN76 A0A1S3SHC9 A0A3B4CUF8 A0A3B3TED3 A0A3Q3KMV7 A0A1S3N0F3 A0A3Q1F4B6 A0A147BU85 A0A131XVU3 W5P3P1 A0A131XF45 W5LKM1 A0A1B6MI28 A0A1B6KHJ9 L7MIH9 I3JUG3
PDB
4U7M
E-value=7.08465e-68,
Score=657
Ontologies
KEGG
GO
Topology
Subcellular location
Cell membrane
Detected in cytoplasmic vesicles when coexpressed with ERBB4. With evidence from 1 publications.
Cytoplasmic vesicle membrane Detected in cytoplasmic vesicles when coexpressed with ERBB4. With evidence from 1 publications.
Cytoplasmic vesicle membrane Detected in cytoplasmic vesicles when coexpressed with ERBB4. With evidence from 1 publications.
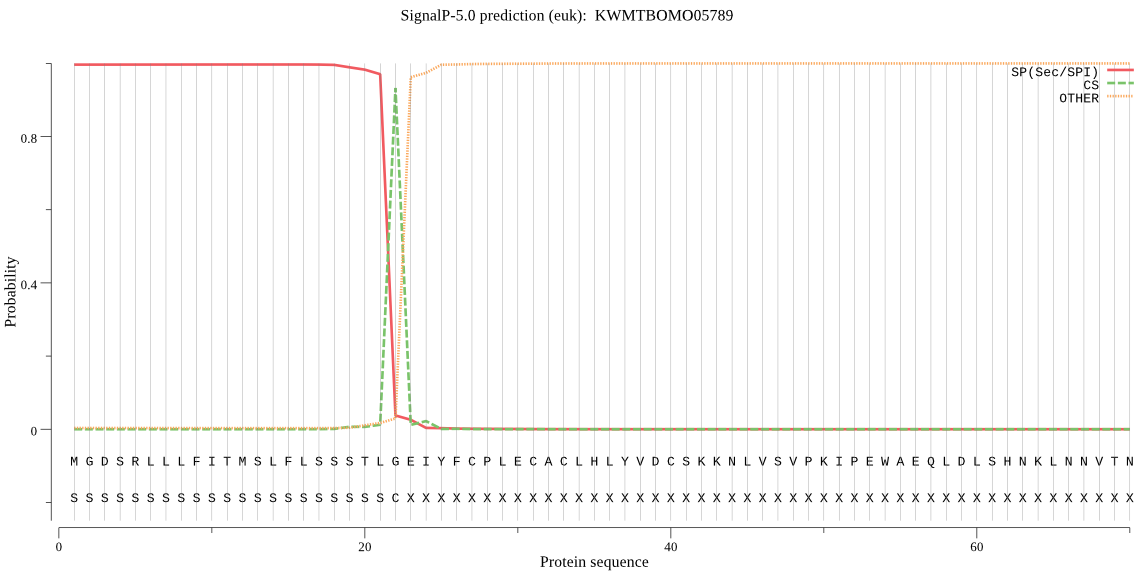
SignalP
Position: 1 - 22,
Likelihood: 0.996707
Length:
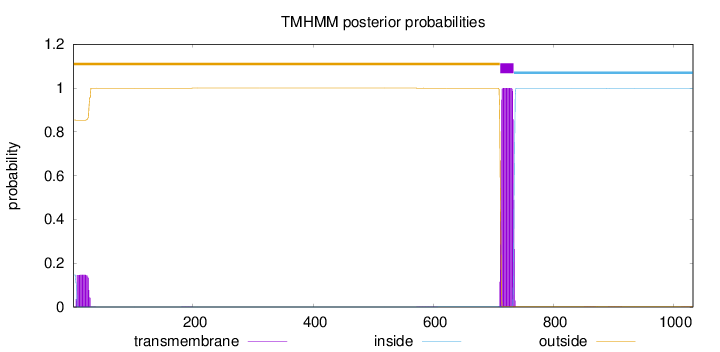
1033
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.95096
Exp number, first 60 AAs:
3.14039
Total prob of N-in:
0.14645
outside
1 - 711
TMhelix
712 - 734
inside
735 - 1033
Population Genetic Test Statistics
Pi
236.425944
Theta
178.691513
Tajima's D
-1.360933
CLR
0.876088
CSRT
0.0776961151942403
Interpretation
Uncertain
