Gene
KWMTBOMO05785
Pre Gene Modal
BGIBMGA006922
Annotation
PREDICTED:_potassium_voltage-gated_channel_subfamily_H_member_8_isoform_X3_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.416
Sequence
CDS
ATGATAGTTCTTAGAACAGAGACACCGCTTCTCAAGAAGGCTGGTTGATCGAGGGCGGGTGCGCGTGGTGCGCGTCGTAAGGGGATGCCCGTGCGCAAGGGGCTTCTCGCGCCCCAGAACACCTTCCTCGATACCATCGCAACTCGTTTCGATGGCACTCACAGTAACTTCGTGCTTGGTAATGCACAAGTGCCGTCGTACCCGATCGTGTACTGCTCAGATGGGTTCTGTGAGCTGACAGGCTGGACAAGGGCCAACATAATGCAAAAGGCCTGCGCCTGTAAGTTCCTCCACGGCCCAGACACGAGAGATGAACATCGCAAGGAGATTGACACCGCACTGGATACCAAACACGAGCTGAAATTGGAGCTTATCTTTTATAAGAAAAATGGATCTCCTTTTTGGTGTCTGCTCGATATCGTTCCAATTAAGAACGAGAAACGAGAAGTCGTTTTATTTCTCGCTTCGTTCAAAGACATCACCAATACCAAGATGGCCGCCATGAACACTAACGAGGAATTCGACAGCGACCCGAATGGGAATCTTGACCCAGAAGCGCCCTCGCCCGCGAACATGGGTAGGCGGCGCTCGCGTGCTGTACTATACCAGCTTTCAGGACATTACAAACCAGACAAAATGAAGACTAAACTCAAACTCAACAATAACCTACTTCACTCAACCGATCCGCCTTTACCAGAATACAAAACGTCCGCTGTGAAAAAGTCGAGATTTATAATATCGCACTATGGGGTTTTTAAAACATTTTGGGACTGGCTCATTCTCATAGCGACATTTTACGTGGCTGTCGTAGTCCCATACAATGCGAGCTTTGTAGACGAAGGCCATCCTAGGATCAGCGTAACGAGTGACGTCATTGTGGAAGCTCTCTTCATTGTCGATATCGTACTCAACTTCCGAACGACGTTCGTGAGCAAGAAAGGAGAAGTAGTTTCGGACTCAAAAGCGATAGCCCTTAATTATATCCGGTCTTGGTTCGTGGTAGATCTCTTGGCGGCGCTCCCGTTTGACCTACTTTACGCTTCGGACGTATACAGCGGAGCGGAATCAACCCACGGAAATGTGCATTTAGTGAAGTTAACGAGGTTGCTACGACTGGCGCGACTCCTGCAGAAGATGGATCGATACTCCCAGTATTCGGCGCTGATATTGACCTTATTGATGCTATCCTTTACGCTTGTCGCGCACTGGCTCGCCTGCATTTGGTTCATAATAGCGGAAAAAGAAATTGAAGTTCACAAAAATGAGATTTGGGATTTAGGCTGGATAAACAACCTAGCGGAGAGACTGAAAGTGCCCATCATTAACATATCACACAGCGAGAGCTACGTCACCGCACTTTACTTCACCTGTTCCTCTCTGACAAGCGTCGGCTTCGGCAACGTTTCGGCGAACACTCTCCCAGAGAAGATTTTCAGCATAATCACCATGTTGATTGGAGCTCTAATGCACGCAGTGGTTTTTGGAAACGTCACGGCGATCATTCAAAGGATGTACTCTCGGAGGTCAATGTACCAGAGCAAGTGGCGTGATCTGAAGGACTTTCTCACTATCAATCAAGTTCCGACGGAGCTGAAACAAAGAATGCAGGATTATTTCCAAACGATGTGGTCGCTAAATCATGGCATCGATATTCATGAGACGCTTAAAGAATTTCCCGAAGAACTGAGAGGCGACGTATCGCTACATTTGCATCGTGAGATCTTGTCACTTCCAATATTCGAATCCGCGTCCCAGGGATGTCTCAAATTGCTTTCGCTACATATTCGGAACAACTTCTGCGCCCCCGGCGAATACCTCGTCCACAAAGGCGATGCCCTCACGTACATCTATTACATCTGCAACGGATCCATGGAGGTGGTACAAAACGATATGGTCGTCGCTATTCTCGGTAAAGGAGATTTAGTTGGGTGCGACGTAAACACCCATTTACAGGCTTACAACGGTACGGGAACGTCTCAAGGAAACAATCCAGACATCGTCGTCAAATCTAGTTGTGATGTAAAGGCTCTGACATACTGCGATTTGAAATGTATTCACATGGCCGGTCTAGCAGAAGTGTTGCGATTGTACCCCGAATACCAGCAAGAGTTTATTCATGACATACAACACGACCTTACTTACAACTTGAGGGAGGGATATGAGGCTGAACAGGAATCAGACGGGAATGGACATCCATCTTTAACGTTACCTTCCATATCAGAAGACGATGAAAACGCTGCGGAAGATCATGCTTTAACACCTAAAAAGCTGCCTTCTTCTAACACTAATAGTCCCCGTCACACTAAATTTAGGATTGACGGACAACGCCTGTCTCACAGGGAGTTACGGGAGAGGATAGAAAGGCAACGCTCTGTCGCAACACCGAAGATAACTCGAGCTGATTCCTTGGAAGGTCTAAATTTAGAAGTGCATAATACGAGGTCTTCTGTTGAAAGACTGGATACACAAGTGTCTACTTTACATCAAGATGTCGCAACACTCAGTATGGAGGTAAGGAATGCCATACAGGCACTTCAAGAAATGACAGGGCCAGCAGGATGGCACGCGGCGCACTCTATACCAAACTTGCAGTGGAACGCGCCCCCCAACCAACTTGCAAGGAGCTGCAGCCATCCACCGGATGTCTTTTGCTGGGATCAACAGGAGAGACCAGTAACTCCTGAGCGCCCGAAAATGCACCGCAGCACGCAAACTGAGCCATTCCTTCACAGCGTAACCCAATATATCATGGAGCATCCAGCTACTGTCATGTTATTACTCGGTATCGATCCGATGTCGAGTTTGCCACCCGTCATTCCGCAGCCCATCGATTATTACGACACTCGACGCCGACCTAGCGCCCTAGAACAGATTATAGAGACTGAAACCGGAAGTCACACCCCTTCTAGTACCACCAGTGAGAAATTCGAACCGATCCGCAGCCTCAGTGGGAGTGCCAAAGAACTGACTTCCCACGATCTAAGGAAGCAATTTGAGCGAGAAAGCAAAGAACGTTTGATACGAAATCGGTATTCGACGAGTGATATACACGAGGAATCGGAACGATTGCTTGGCGCGAACAGGACACATTCAAGCACCCGTAGCCTCAAGTACAACATTAACAGTTAA
Protein
MIVLRTETPLLKKAGXSRAGARGARRKGMPVRKGLLAPQNTFLDTIATRFDGTHSNFVLGNAQVPSYPIVYCSDGFCELTGWTRANIMQKACACKFLHGPDTRDEHRKEIDTALDTKHELKLELIFYKKNGSPFWCLLDIVPIKNEKREVVLFLASFKDITNTKMAAMNTNEEFDSDPNGNLDPEAPSPANMGRRRSRAVLYQLSGHYKPDKMKTKLKLNNNLLHSTDPPLPEYKTSAVKKSRFIISHYGVFKTFWDWLILIATFYVAVVVPYNASFVDEGHPRISVTSDVIVEALFIVDIVLNFRTTFVSKKGEVVSDSKAIALNYIRSWFVVDLLAALPFDLLYASDVYSGAESTHGNVHLVKLTRLLRLARLLQKMDRYSQYSALILTLLMLSFTLVAHWLACIWFIIAEKEIEVHKNEIWDLGWINNLAERLKVPIINISHSESYVTALYFTCSSLTSVGFGNVSANTLPEKIFSIITMLIGALMHAVVFGNVTAIIQRMYSRRSMYQSKWRDLKDFLTINQVPTELKQRMQDYFQTMWSLNHGIDIHETLKEFPEELRGDVSLHLHREILSLPIFESASQGCLKLLSLHIRNNFCAPGEYLVHKGDALTYIYYICNGSMEVVQNDMVVAILGKGDLVGCDVNTHLQAYNGTGTSQGNNPDIVVKSSCDVKALTYCDLKCIHMAGLAEVLRLYPEYQQEFIHDIQHDLTYNLREGYEAEQESDGNGHPSLTLPSISEDDENAAEDHALTPKKLPSSNTNSPRHTKFRIDGQRLSHRELRERIERQRSVATPKITRADSLEGLNLEVHNTRSSVERLDTQVSTLHQDVATLSMEVRNAIQALQEMTGPAGWHAAHSIPNLQWNAPPNQLARSCSHPPDVFCWDQQERPVTPERPKMHRSTQTEPFLHSVTQYIMEHPATVMLLLGIDPMSSLPPVIPQPIDYYDTRRRPSALEQIIETETGSHTPSSTTSEKFEPIRSLSGSAKELTSHDLRKQFERESKERLIRNRYSTSDIHEESERLLGANRTHSSTRSLKYNINS
Summary
Uniprot
A0A2A4K0Y2
A0A2A4K1N3
A0A2A4K208
A0A2W1C0C8
A0A194Q8R8
A0A212ENU5
+ More
A0A2H1W4R6 A0A194QQP7 A0A1B6DT39 A0A3L8DC27 A0A088ASQ3 D6X119 A0A195DPF0 A0A0L7R6A8 A0A195D8D0 A0A1L8DDH7 A0A1L8DDU7 A0A182J9T5 A0A151WPF5 A0A182N9M7 A0A182LGS0 A0A026WWJ1 A0A182FCT8 Q7Q9F5 A0A182H5E7 A0A182RII8 A0A158NFQ5 A0A0J7L0L1 A0A182PVF6 A0A067R7J4 A0A139WCK3 A0A2S2PZI2 A0A2S2PMW9 A0A2S2PDF3 W8CAA9 A0A154PF88 A0A3B0JR24 A0A195BDL1 A0A151JYL6 B4P513 A0A0R3NM41 A0A0R3NSM1 A0A0R1DS68 B4LJZ2 Q290R8 A0A1I8NWK3 B4GBC0 A0A0J9RFM1 A0A0J9RFG2 B4J6H9 A0A0J9RF72 A0A0J9U4Y3 A0A0J9RFM5 B3NLM2 A0A0J9RFG7 B4HN99 A0A0B4LGW2 B3MHW3 B4KNK1 A0A1W4VD11 C7LA62 A1ZB14 Q23974 A0A1W4V0A8 B4MYK1 A0A1W4VCK4 E9ITH2 A0A182HNT3 A0A182X8I1 A0A1C9TA84 A0A0M3QVC1 A0A1C9TAB3 J9L063 A0A182QHL3 B0WTC0 A0A1S4JS72 Q0IEW9 A0A1S3I8E8 A0A0A9W8E5 A0A3S3PK05 A0A1L8DDG3 A0A1L8DET2 A0A0R3NMC9 A0A0J9RGS3 C4IXY2 A0A2S1WM35 A0A1L8EPM4 I3JA06 A0A1W4ZZ20
A0A2H1W4R6 A0A194QQP7 A0A1B6DT39 A0A3L8DC27 A0A088ASQ3 D6X119 A0A195DPF0 A0A0L7R6A8 A0A195D8D0 A0A1L8DDH7 A0A1L8DDU7 A0A182J9T5 A0A151WPF5 A0A182N9M7 A0A182LGS0 A0A026WWJ1 A0A182FCT8 Q7Q9F5 A0A182H5E7 A0A182RII8 A0A158NFQ5 A0A0J7L0L1 A0A182PVF6 A0A067R7J4 A0A139WCK3 A0A2S2PZI2 A0A2S2PMW9 A0A2S2PDF3 W8CAA9 A0A154PF88 A0A3B0JR24 A0A195BDL1 A0A151JYL6 B4P513 A0A0R3NM41 A0A0R3NSM1 A0A0R1DS68 B4LJZ2 Q290R8 A0A1I8NWK3 B4GBC0 A0A0J9RFM1 A0A0J9RFG2 B4J6H9 A0A0J9RF72 A0A0J9U4Y3 A0A0J9RFM5 B3NLM2 A0A0J9RFG7 B4HN99 A0A0B4LGW2 B3MHW3 B4KNK1 A0A1W4VD11 C7LA62 A1ZB14 Q23974 A0A1W4V0A8 B4MYK1 A0A1W4VCK4 E9ITH2 A0A182HNT3 A0A182X8I1 A0A1C9TA84 A0A0M3QVC1 A0A1C9TAB3 J9L063 A0A182QHL3 B0WTC0 A0A1S4JS72 Q0IEW9 A0A1S3I8E8 A0A0A9W8E5 A0A3S3PK05 A0A1L8DDG3 A0A1L8DET2 A0A0R3NMC9 A0A0J9RGS3 C4IXY2 A0A2S1WM35 A0A1L8EPM4 I3JA06 A0A1W4ZZ20
Pubmed
28756777
26354079
22118469
30249741
18362917
19820115
+ More
20966253 24508170 12364791 14747013 17210077 26483478 21347285 24845553 24495485 17994087 17550304 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23555008 8159766 21282665 17510324 25401762 27762356 25186727
20966253 24508170 12364791 14747013 17210077 26483478 21347285 24845553 24495485 17994087 17550304 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23555008 8159766 21282665 17510324 25401762 27762356 25186727
EMBL
NWSH01000259
PCG77935.1
PCG77936.1
PCG77934.1
KZ149900
PZC78570.1
+ More
KQ459302 KPJ01794.1 AGBW02013613 OWR43172.1 ODYU01006193 SOQ47832.1 KQ461175 KPJ07822.1 GEDC01008465 JAS28833.1 QOIP01000010 RLU17891.1 KQ971367 EFA09395.2 KQ980662 KYN14687.1 KQ414646 KOC66422.1 KQ976750 KYN08699.1 GFDF01009573 JAV04511.1 GFDF01009574 JAV04510.1 KQ982893 KYQ49575.1 KK107079 EZA60183.1 AAAB01008900 EAA09546.4 JXUM01004210 JXUM01004211 JXUM01004212 JXUM01004213 JXUM01004214 JXUM01004215 KQ560171 KXJ84016.1 ADTU01014532 ADTU01014533 ADTU01014534 ADTU01014535 LBMM01001544 KMQ96113.1 KK852824 KDR15465.1 KYB25650.1 GGMS01001636 MBY70839.1 GGMR01018136 MBY30755.1 GGMR01014803 MBY27422.1 GAMC01005926 JAC00630.1 KQ434890 KZC10472.1 OUUW01000001 SPP73598.1 KQ976514 KYM82299.1 KQ981494 KYN40954.1 CM000158 EDW91714.2 CM000071 KRT02046.1 KRT02044.1 KRT02045.1 KRK00044.1 CH940648 EDW61646.1 EAL25294.3 CH479181 EDW32222.1 CM002911 KMY94706.1 KMY94707.1 CH916367 EDW01980.1 KMY94708.1 KMY94710.1 KMY94711.1 CH954179 EDV54997.1 KMY94712.1 CH480816 EDW48381.1 AE013599 AHN56349.1 CH902619 EDV36950.1 CH933808 EDW08960.2 BT099612 ACU51756.1 AAF57772.2 U04246 AAA62472.1 CH963894 EDW77190.1 GL765555 EFZ16129.1 APCN01002763 KU681447 AOR07228.1 CP012524 ALC42159.1 KU681459 AOR07240.1 ABLF02023417 ABLF02023418 ABLF02023419 ABLF02023420 ABLF02023421 ABLF02023422 ABLF02046863 ABLF02057972 ABLF02059504 ABLF02066165 AXCN02001012 DS232084 EDS34354.1 CH477464 EAT40502.1 GBHO01042514 JAG01090.1 NCKU01000814 RWS14048.1 GFDF01009592 JAV04492.1 GFDF01009128 JAV04956.1 KRT02043.1 KMY94709.1 BT083413 ACQ89822.1 MG973395 AWJ68248.1 CM004483 OCT61292.1 AERX01020407 AERX01020408 AERX01020409
KQ459302 KPJ01794.1 AGBW02013613 OWR43172.1 ODYU01006193 SOQ47832.1 KQ461175 KPJ07822.1 GEDC01008465 JAS28833.1 QOIP01000010 RLU17891.1 KQ971367 EFA09395.2 KQ980662 KYN14687.1 KQ414646 KOC66422.1 KQ976750 KYN08699.1 GFDF01009573 JAV04511.1 GFDF01009574 JAV04510.1 KQ982893 KYQ49575.1 KK107079 EZA60183.1 AAAB01008900 EAA09546.4 JXUM01004210 JXUM01004211 JXUM01004212 JXUM01004213 JXUM01004214 JXUM01004215 KQ560171 KXJ84016.1 ADTU01014532 ADTU01014533 ADTU01014534 ADTU01014535 LBMM01001544 KMQ96113.1 KK852824 KDR15465.1 KYB25650.1 GGMS01001636 MBY70839.1 GGMR01018136 MBY30755.1 GGMR01014803 MBY27422.1 GAMC01005926 JAC00630.1 KQ434890 KZC10472.1 OUUW01000001 SPP73598.1 KQ976514 KYM82299.1 KQ981494 KYN40954.1 CM000158 EDW91714.2 CM000071 KRT02046.1 KRT02044.1 KRT02045.1 KRK00044.1 CH940648 EDW61646.1 EAL25294.3 CH479181 EDW32222.1 CM002911 KMY94706.1 KMY94707.1 CH916367 EDW01980.1 KMY94708.1 KMY94710.1 KMY94711.1 CH954179 EDV54997.1 KMY94712.1 CH480816 EDW48381.1 AE013599 AHN56349.1 CH902619 EDV36950.1 CH933808 EDW08960.2 BT099612 ACU51756.1 AAF57772.2 U04246 AAA62472.1 CH963894 EDW77190.1 GL765555 EFZ16129.1 APCN01002763 KU681447 AOR07228.1 CP012524 ALC42159.1 KU681459 AOR07240.1 ABLF02023417 ABLF02023418 ABLF02023419 ABLF02023420 ABLF02023421 ABLF02023422 ABLF02046863 ABLF02057972 ABLF02059504 ABLF02066165 AXCN02001012 DS232084 EDS34354.1 CH477464 EAT40502.1 GBHO01042514 JAG01090.1 NCKU01000814 RWS14048.1 GFDF01009592 JAV04492.1 GFDF01009128 JAV04956.1 KRT02043.1 KMY94709.1 BT083413 ACQ89822.1 MG973395 AWJ68248.1 CM004483 OCT61292.1 AERX01020407 AERX01020408 AERX01020409
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000279307
UP000005203
+ More
UP000007266 UP000078492 UP000053825 UP000078542 UP000075880 UP000075809 UP000075884 UP000075882 UP000053097 UP000069272 UP000007062 UP000069940 UP000249989 UP000075900 UP000005205 UP000036403 UP000075885 UP000027135 UP000076502 UP000268350 UP000078540 UP000078541 UP000002282 UP000001819 UP000008792 UP000095300 UP000008744 UP000001070 UP000008711 UP000001292 UP000000803 UP000007801 UP000009192 UP000192221 UP000007798 UP000075840 UP000076407 UP000092553 UP000007819 UP000075886 UP000002320 UP000008820 UP000085678 UP000285301 UP000186698 UP000005207 UP000192224
UP000007266 UP000078492 UP000053825 UP000078542 UP000075880 UP000075809 UP000075884 UP000075882 UP000053097 UP000069272 UP000007062 UP000069940 UP000249989 UP000075900 UP000005205 UP000036403 UP000075885 UP000027135 UP000076502 UP000268350 UP000078540 UP000078541 UP000002282 UP000001819 UP000008792 UP000095300 UP000008744 UP000001070 UP000008711 UP000001292 UP000000803 UP000007801 UP000009192 UP000192221 UP000007798 UP000075840 UP000076407 UP000092553 UP000007819 UP000075886 UP000002320 UP000008820 UP000085678 UP000285301 UP000186698 UP000005207 UP000192224
Interpro
IPR014710
RmlC-like_jellyroll
+ More
IPR018490 cNMP-bd-like
IPR035965 PAS-like_dom_sf
IPR001610 PAC
IPR000014 PAS
IPR027359 Volt_channel_dom_sf
IPR003938 K_chnl_volt-dep_EAG/ELK/ERG
IPR000595 cNMP-bd_dom
IPR005821 Ion_trans_dom
IPR003950 K_chnl_volt-dep_ELK
IPR000700 PAS-assoc_C
IPR035897 Toll_tir_struct_dom_sf
IPR000157 TIR_dom
IPR003967 K_chnl_volt-dep_ERG
IPR016184 Capsid/spike_ssDNA_virus
IPR003433 Capsid_VP4_densovirus
IPR018490 cNMP-bd-like
IPR035965 PAS-like_dom_sf
IPR001610 PAC
IPR000014 PAS
IPR027359 Volt_channel_dom_sf
IPR003938 K_chnl_volt-dep_EAG/ELK/ERG
IPR000595 cNMP-bd_dom
IPR005821 Ion_trans_dom
IPR003950 K_chnl_volt-dep_ELK
IPR000700 PAS-assoc_C
IPR035897 Toll_tir_struct_dom_sf
IPR000157 TIR_dom
IPR003967 K_chnl_volt-dep_ERG
IPR016184 Capsid/spike_ssDNA_virus
IPR003433 Capsid_VP4_densovirus
Gene 3D
ProteinModelPortal
A0A2A4K0Y2
A0A2A4K1N3
A0A2A4K208
A0A2W1C0C8
A0A194Q8R8
A0A212ENU5
+ More
A0A2H1W4R6 A0A194QQP7 A0A1B6DT39 A0A3L8DC27 A0A088ASQ3 D6X119 A0A195DPF0 A0A0L7R6A8 A0A195D8D0 A0A1L8DDH7 A0A1L8DDU7 A0A182J9T5 A0A151WPF5 A0A182N9M7 A0A182LGS0 A0A026WWJ1 A0A182FCT8 Q7Q9F5 A0A182H5E7 A0A182RII8 A0A158NFQ5 A0A0J7L0L1 A0A182PVF6 A0A067R7J4 A0A139WCK3 A0A2S2PZI2 A0A2S2PMW9 A0A2S2PDF3 W8CAA9 A0A154PF88 A0A3B0JR24 A0A195BDL1 A0A151JYL6 B4P513 A0A0R3NM41 A0A0R3NSM1 A0A0R1DS68 B4LJZ2 Q290R8 A0A1I8NWK3 B4GBC0 A0A0J9RFM1 A0A0J9RFG2 B4J6H9 A0A0J9RF72 A0A0J9U4Y3 A0A0J9RFM5 B3NLM2 A0A0J9RFG7 B4HN99 A0A0B4LGW2 B3MHW3 B4KNK1 A0A1W4VD11 C7LA62 A1ZB14 Q23974 A0A1W4V0A8 B4MYK1 A0A1W4VCK4 E9ITH2 A0A182HNT3 A0A182X8I1 A0A1C9TA84 A0A0M3QVC1 A0A1C9TAB3 J9L063 A0A182QHL3 B0WTC0 A0A1S4JS72 Q0IEW9 A0A1S3I8E8 A0A0A9W8E5 A0A3S3PK05 A0A1L8DDG3 A0A1L8DET2 A0A0R3NMC9 A0A0J9RGS3 C4IXY2 A0A2S1WM35 A0A1L8EPM4 I3JA06 A0A1W4ZZ20
A0A2H1W4R6 A0A194QQP7 A0A1B6DT39 A0A3L8DC27 A0A088ASQ3 D6X119 A0A195DPF0 A0A0L7R6A8 A0A195D8D0 A0A1L8DDH7 A0A1L8DDU7 A0A182J9T5 A0A151WPF5 A0A182N9M7 A0A182LGS0 A0A026WWJ1 A0A182FCT8 Q7Q9F5 A0A182H5E7 A0A182RII8 A0A158NFQ5 A0A0J7L0L1 A0A182PVF6 A0A067R7J4 A0A139WCK3 A0A2S2PZI2 A0A2S2PMW9 A0A2S2PDF3 W8CAA9 A0A154PF88 A0A3B0JR24 A0A195BDL1 A0A151JYL6 B4P513 A0A0R3NM41 A0A0R3NSM1 A0A0R1DS68 B4LJZ2 Q290R8 A0A1I8NWK3 B4GBC0 A0A0J9RFM1 A0A0J9RFG2 B4J6H9 A0A0J9RF72 A0A0J9U4Y3 A0A0J9RFM5 B3NLM2 A0A0J9RFG7 B4HN99 A0A0B4LGW2 B3MHW3 B4KNK1 A0A1W4VD11 C7LA62 A1ZB14 Q23974 A0A1W4V0A8 B4MYK1 A0A1W4VCK4 E9ITH2 A0A182HNT3 A0A182X8I1 A0A1C9TA84 A0A0M3QVC1 A0A1C9TAB3 J9L063 A0A182QHL3 B0WTC0 A0A1S4JS72 Q0IEW9 A0A1S3I8E8 A0A0A9W8E5 A0A3S3PK05 A0A1L8DDG3 A0A1L8DET2 A0A0R3NMC9 A0A0J9RGS3 C4IXY2 A0A2S1WM35 A0A1L8EPM4 I3JA06 A0A1W4ZZ20
PDB
5VA2
E-value=1.87051e-148,
Score=1352
Ontologies
GO
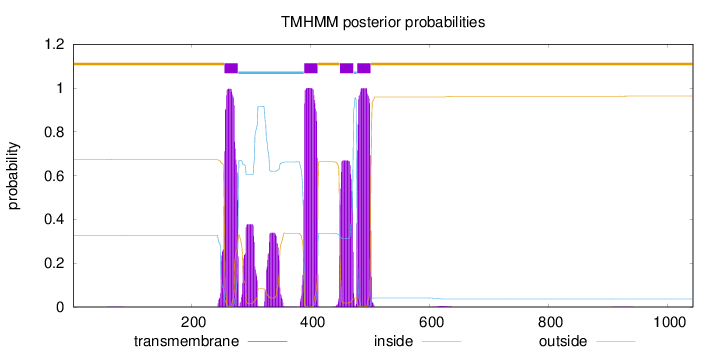
Topology
Length:
1042
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
97.90391
Exp number, first 60 AAs:
0.00126
Total prob of N-in:
0.32658
outside
1 - 254
TMhelix
255 - 277
inside
278 - 388
TMhelix
389 - 411
outside
412 - 448
TMhelix
449 - 471
inside
472 - 477
TMhelix
478 - 500
outside
501 - 1042
Population Genetic Test Statistics
Pi
220.301808
Theta
178.863563
Tajima's D
0.764707
CLR
0.454317
CSRT
0.589370531473426
Interpretation
Uncertain
