Pre Gene Modal
BGIBMGA006932
Annotation
PREDICTED:_NECAP-like_protein_CG9132_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.914
Sequence
CDS
ATGGATACTTATGAAAGTGTTATTTTAGTGAAGAACGAAGTATTCGTTTTTAAAATACCACCCAGAACATCTAACAGAGGATACAGGGCAGCAGATTGGAACTTGCAAGAGCCACAATGGACAGGTCGCATGAGGTTGGTGTCAAAAGGGAACGAGTTGGTCATGAAGTTAGAGGATAAAACAAGTGGCGAATTATTTGCGAAGTGCCCAATAGATAAATATCCAGGAGTTGCACTGGAAGCTGTAACTGATAGCTCACGATATTTTGTTGTGAAAATACAAGATGATAATGGTTGTGCTGCATATATCGGTCTTGGATTTGGTGATAGATCTGATTCGTTTGATCTAAATGTTGCACTTCAAGACCATTTCAAATGGTTGAAAAAAGAACAAGAGAGCGATCAAACACCACAGGGACAATTGGATCTTGGATTTAAAGATGGTGAAACAATTAAAATCAACATGAAAATTACTAAAAAAGATGGGAGTGAGGGCAGCAGGTCACGCCGTGGTGTGCCAGGTGCTAGTGGTGGTGGTTTGTTGCCACCACCTCCAGGCGGTTCTAAGTTACCACCTCCACCATCACCACAACATGCTGCTTCACCATCATCAATCCCACCTGCCAGCTCTAACACAGAGTGGGGTGAATTTAATTCAGCAAGTGCTCCAGAACCACAAGCAACAACCCCAGCCAATCAACAAAATTGGGTGCAGTTCTGA
Protein
MDTYESVILVKNEVFVFKIPPRTSNRGYRAADWNLQEPQWTGRMRLVSKGNELVMKLEDKTSGELFAKCPIDKYPGVALEAVTDSSRYFVVKIQDDNGCAAYIGLGFGDRSDSFDLNVALQDHFKWLKKEQESDQTPQGQLDLGFKDGETIKINMKITKKDGSEGSRSRRGVPGASGGGLLPPPPGGSKLPPPPSPQHAASPSSIPPASSNTEWGEFNSASAPEPQATTPANQQNWVQF
Summary
Uniprot
A0A2A4JNP0
A0A2H1W7X8
A0A2W1BW55
A0A3S2MAM7
A0A194Q9X7
A0A194RGD6
+ More
S4NPX1 A0A212ENZ0 A0A1Y1KPH9 D6X1A3 B0XD83 A0A1Q3EZA2 A0A1Q3EZF0 A0A1Q3EZB2 A0A182GPJ2 T1I3S8 A0A1B0CHK8 A0A0L7QNP9 A0A0V0G5K2 A0A224XRM0 A0A023F439 A0A069DR24 A0A1L8DAU9 A0A2A3EDW9 A0A088A7I9 A0A026WT72 A0A0A9WV73 U5EZQ1 A0A0K8TNX5 E2BXA6 A0A1B0DNM5 A0A182JFS4 A0A0P5E8W3 A0A146LY01 A0A084VJF4 A0A0P4XPH5 A0A0P5U6W0 A0A2M4BX58 A0A0P5J5P3 A0A0P5ZJR5 W5JNR6 A0A0T6AWX8 A0A182FB23 A0A0P6C4X2 A0A164WP81
S4NPX1 A0A212ENZ0 A0A1Y1KPH9 D6X1A3 B0XD83 A0A1Q3EZA2 A0A1Q3EZF0 A0A1Q3EZB2 A0A182GPJ2 T1I3S8 A0A1B0CHK8 A0A0L7QNP9 A0A0V0G5K2 A0A224XRM0 A0A023F439 A0A069DR24 A0A1L8DAU9 A0A2A3EDW9 A0A088A7I9 A0A026WT72 A0A0A9WV73 U5EZQ1 A0A0K8TNX5 E2BXA6 A0A1B0DNM5 A0A182JFS4 A0A0P5E8W3 A0A146LY01 A0A084VJF4 A0A0P4XPH5 A0A0P5U6W0 A0A2M4BX58 A0A0P5J5P3 A0A0P5ZJR5 W5JNR6 A0A0T6AWX8 A0A182FB23 A0A0P6C4X2 A0A164WP81
Pubmed
EMBL
NWSH01000905
PCG73591.1
ODYU01006887
SOQ49133.1
KZ149900
PZC78571.1
+ More
RSAL01000003 RVE54872.1 KQ459302 KPJ01795.1 KQ460398 KPJ14996.1 GAIX01013411 JAA79149.1 AGBW02013613 OWR43171.1 GEZM01082507 JAV61346.1 KQ971372 EFA09517.2 DS232745 EDS45337.1 GFDL01014415 JAV20630.1 GFDL01014369 JAV20676.1 GFDL01014403 JAV20642.1 JXUM01015121 JXUM01015122 JXUM01015123 KQ560423 KXJ82483.1 ACPB03021980 AJWK01012445 AJWK01012446 AJWK01012447 KQ414851 KOC60194.1 GECL01002869 JAP03255.1 GFTR01003958 JAW12468.1 GBBI01002481 JAC16231.1 GBGD01002728 JAC86161.1 GFDF01010483 JAV03601.1 KZ288279 PBC29674.1 KK107109 QOIP01000008 EZA59138.1 RLU19797.1 GBHO01035659 GBHO01035658 GBHO01032005 GBHO01032000 GBHO01002971 GDHC01006408 GDHC01003137 GDHC01000216 JAG07945.1 JAG07946.1 JAG11599.1 JAG11604.1 JAG40633.1 JAQ12221.1 JAQ15492.1 JAQ18413.1 GANO01000374 JAB59497.1 GDAI01001767 JAI15836.1 GL451230 EFN79689.1 AJVK01001445 GDIP01145062 JAJ78340.1 GDHC01007122 JAQ11507.1 ATLV01013556 KE524867 KFB38098.1 GDIP01238252 JAI85149.1 GDIP01118764 JAL84950.1 GGFJ01008516 MBW57657.1 GDIQ01210666 GDIQ01121543 JAK41059.1 GDIP01042977 JAM60738.1 ADMH02000974 ETN64540.1 LJIG01022648 KRT79451.1 GDIP01009138 JAM94577.1 LRGB01001151 KZS13450.1
RSAL01000003 RVE54872.1 KQ459302 KPJ01795.1 KQ460398 KPJ14996.1 GAIX01013411 JAA79149.1 AGBW02013613 OWR43171.1 GEZM01082507 JAV61346.1 KQ971372 EFA09517.2 DS232745 EDS45337.1 GFDL01014415 JAV20630.1 GFDL01014369 JAV20676.1 GFDL01014403 JAV20642.1 JXUM01015121 JXUM01015122 JXUM01015123 KQ560423 KXJ82483.1 ACPB03021980 AJWK01012445 AJWK01012446 AJWK01012447 KQ414851 KOC60194.1 GECL01002869 JAP03255.1 GFTR01003958 JAW12468.1 GBBI01002481 JAC16231.1 GBGD01002728 JAC86161.1 GFDF01010483 JAV03601.1 KZ288279 PBC29674.1 KK107109 QOIP01000008 EZA59138.1 RLU19797.1 GBHO01035659 GBHO01035658 GBHO01032005 GBHO01032000 GBHO01002971 GDHC01006408 GDHC01003137 GDHC01000216 JAG07945.1 JAG07946.1 JAG11599.1 JAG11604.1 JAG40633.1 JAQ12221.1 JAQ15492.1 JAQ18413.1 GANO01000374 JAB59497.1 GDAI01001767 JAI15836.1 GL451230 EFN79689.1 AJVK01001445 GDIP01145062 JAJ78340.1 GDHC01007122 JAQ11507.1 ATLV01013556 KE524867 KFB38098.1 GDIP01238252 JAI85149.1 GDIP01118764 JAL84950.1 GGFJ01008516 MBW57657.1 GDIQ01210666 GDIQ01121543 JAK41059.1 GDIP01042977 JAM60738.1 ADMH02000974 ETN64540.1 LJIG01022648 KRT79451.1 GDIP01009138 JAM94577.1 LRGB01001151 KZS13450.1
Proteomes
Pfam
PF07933 DUF1681
Gene 3D
ProteinModelPortal
A0A2A4JNP0
A0A2H1W7X8
A0A2W1BW55
A0A3S2MAM7
A0A194Q9X7
A0A194RGD6
+ More
S4NPX1 A0A212ENZ0 A0A1Y1KPH9 D6X1A3 B0XD83 A0A1Q3EZA2 A0A1Q3EZF0 A0A1Q3EZB2 A0A182GPJ2 T1I3S8 A0A1B0CHK8 A0A0L7QNP9 A0A0V0G5K2 A0A224XRM0 A0A023F439 A0A069DR24 A0A1L8DAU9 A0A2A3EDW9 A0A088A7I9 A0A026WT72 A0A0A9WV73 U5EZQ1 A0A0K8TNX5 E2BXA6 A0A1B0DNM5 A0A182JFS4 A0A0P5E8W3 A0A146LY01 A0A084VJF4 A0A0P4XPH5 A0A0P5U6W0 A0A2M4BX58 A0A0P5J5P3 A0A0P5ZJR5 W5JNR6 A0A0T6AWX8 A0A182FB23 A0A0P6C4X2 A0A164WP81
S4NPX1 A0A212ENZ0 A0A1Y1KPH9 D6X1A3 B0XD83 A0A1Q3EZA2 A0A1Q3EZF0 A0A1Q3EZB2 A0A182GPJ2 T1I3S8 A0A1B0CHK8 A0A0L7QNP9 A0A0V0G5K2 A0A224XRM0 A0A023F439 A0A069DR24 A0A1L8DAU9 A0A2A3EDW9 A0A088A7I9 A0A026WT72 A0A0A9WV73 U5EZQ1 A0A0K8TNX5 E2BXA6 A0A1B0DNM5 A0A182JFS4 A0A0P5E8W3 A0A146LY01 A0A084VJF4 A0A0P4XPH5 A0A0P5U6W0 A0A2M4BX58 A0A0P5J5P3 A0A0P5ZJR5 W5JNR6 A0A0T6AWX8 A0A182FB23 A0A0P6C4X2 A0A164WP81
PDB
1TQZ
E-value=3.27844e-47,
Score=472
Ontologies
GO
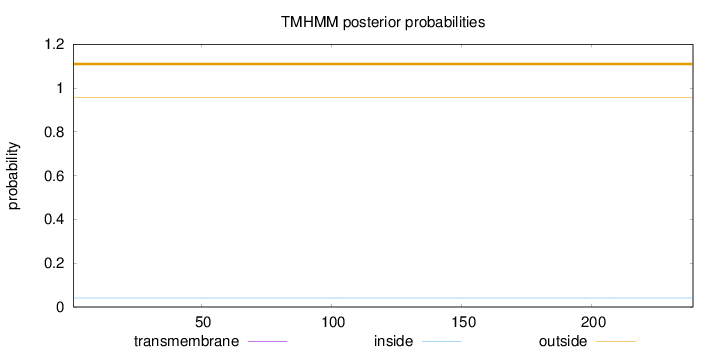
Topology
Length:
239
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00019
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.04163
outside
1 - 239
Population Genetic Test Statistics
Pi
152.015608
Theta
145.683626
Tajima's D
0.469336
CLR
0.014617
CSRT
0.499225038748063
Interpretation
Uncertain
