Gene
KWMTBOMO05781
Pre Gene Modal
BGIBMGA006925
Annotation
PREDICTED:_trafficking_kinesin-binding_protein_milt_isoform_X5_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.136
Sequence
CDS
ATGACAAAAACCTACAATGACATCGAAGCGGTAACACGGCTCTTAGAAGAAAAAGAGAAAGATTTGGAATTGACGGCACGTATCGGTAAGGAATTGCTGACTGCTAATGGACGTTTGGAGTCTCGCGTCACAGCACTTGAAGGAGAGTTGAAAACGGCACGGGACCACATTACGCAACTGAAGCACGATCTCAGTGCCAAGACTGATCTATTACAGGTGCTAACAAATGACACAGAAGAGGGGTCCCCTACGGAGCAGGAGGAGGCGAGTGCGGCTGCGTTGCGACGTCGGACCGGCGCACTGGAGCGGGAGAACCGCGCCCTGCGCGATGAGGCGGCGCGCCTCGCGGCCGGCGCCGACTCCGCCGAGCTCGCCGAGAGACAGCTGCTGCGTGACATCGTGCAACAACTCACAAGTGCGAACTCTGAGGCCAGCGCGTTGGAAACCGAGCTGCTCGAAGAGAGGCAACGCTCTGTCGATCTTCAACAGCAGCTGGACAGCACTCGATCGAGGTTCGCCTTCAACGAAAGAGACTTTAAACAGCTAACGACGGAACATGAGCACACGCTGCGAATATTAGAAATAACAAAGGAGAATCAGAATGCACTCGCAGCGGAACTGGCCGATGTTAAGGAGAGCCATTTCATGTTTTAA
Protein
MTKTYNDIEAVTRLLEEKEKDLELTARIGKELLTANGRLESRVTALEGELKTARDHITQLKHDLSAKTDLLQVLTNDTEEGSPTEQEEASAAALRRRTGALERENRALRDEAARLAAGADSAELAERQLLRDIVQQLTSANSEASALETELLEERQRSVDLQQQLDSTRSRFAFNERDFKQLTTEHEHTLRILEITKENQNALAAELADVKESHFMF
Summary
Uniprot
H9JBN0
A0A2A4JVR8
A0A2A4JV16
A0A2W1C0T8
A0A2H1X0N8
A0A194RGE1
+ More
S4PNM5 A0A3S2NMN8 A0A194Q8D9 A0A1E1VYT7 A0A1E1W9R8 A0A1E1VZA2 A0A1E1W4A5 A0A0L7LU51 A0A0C9R5V9 E0V9Z5 A0A2J7Q8K9 A0A195EXN5 A0A026VZC9 A0A3L8DQB9 E2A3Z3 A0A151JS49 A0A0J7KWE0 E2BK45 A0A151X0D9 A0A158NJ70 A0A195BL26 E9J1U1 A0A151I9Y5 V9I9X8 A0A088AJM3 V9I9F6 A0A232EPM6 A0A023EZK9 U5EUT9 A0A1B6CHY3 A0A2P8YR13 A0A224XSA3 A0A1B6EG82 A0A034V6V3 A0A034V811 A0A0V0G9V9 A0A034V4P0 B4GK01 A0A0R3NXJ6 B5DHM1 A0A0M8ZQ00 A0A0R3NWU7 A0A0R3NR62 A0A034V9L1 A0A1B6GZ72 A0A1B6EZ88 A0A1I8Q555 A0A1B6EWT1 A0A1I8Q583 A0A1B6EZ74 A0A1I8Q549 A0A0K8WCF1 A0A3B0K757 V5GIH2 A0A0K8VVH9 X2D8A5 A0A0A1X155 A0A0A1X7V9 T1PDA2 A0A1Y1NC82 N6SYN6 U4UFN3 A0A1I8MTF7 A0A1I8MTF6 A0A336LLR6 A0A1I8MTF9 A0A336LMH3 A0A0A9Z4M0 A0A0A9Z8Z9 A0A2M4ACF5 A0A2M4ACC6 A0A2M4BBZ3 A0A0A9Z4L5 A0A182FNC4 A0A0A9Z323 A0A0P5UMR5 A0A2M4ABD1 A0A2M4AAM1 A0A0P4YH45 A0A0P5ASR5 W8B3J4 A0A0P5BUA7 A0A0P5B5V2 A0A182H0V2 A0A1Q3F7G3
S4PNM5 A0A3S2NMN8 A0A194Q8D9 A0A1E1VYT7 A0A1E1W9R8 A0A1E1VZA2 A0A1E1W4A5 A0A0L7LU51 A0A0C9R5V9 E0V9Z5 A0A2J7Q8K9 A0A195EXN5 A0A026VZC9 A0A3L8DQB9 E2A3Z3 A0A151JS49 A0A0J7KWE0 E2BK45 A0A151X0D9 A0A158NJ70 A0A195BL26 E9J1U1 A0A151I9Y5 V9I9X8 A0A088AJM3 V9I9F6 A0A232EPM6 A0A023EZK9 U5EUT9 A0A1B6CHY3 A0A2P8YR13 A0A224XSA3 A0A1B6EG82 A0A034V6V3 A0A034V811 A0A0V0G9V9 A0A034V4P0 B4GK01 A0A0R3NXJ6 B5DHM1 A0A0M8ZQ00 A0A0R3NWU7 A0A0R3NR62 A0A034V9L1 A0A1B6GZ72 A0A1B6EZ88 A0A1I8Q555 A0A1B6EWT1 A0A1I8Q583 A0A1B6EZ74 A0A1I8Q549 A0A0K8WCF1 A0A3B0K757 V5GIH2 A0A0K8VVH9 X2D8A5 A0A0A1X155 A0A0A1X7V9 T1PDA2 A0A1Y1NC82 N6SYN6 U4UFN3 A0A1I8MTF7 A0A1I8MTF6 A0A336LLR6 A0A1I8MTF9 A0A336LMH3 A0A0A9Z4M0 A0A0A9Z8Z9 A0A2M4ACF5 A0A2M4ACC6 A0A2M4BBZ3 A0A0A9Z4L5 A0A182FNC4 A0A0A9Z323 A0A0P5UMR5 A0A2M4ABD1 A0A2M4AAM1 A0A0P4YH45 A0A0P5ASR5 W8B3J4 A0A0P5BUA7 A0A0P5B5V2 A0A182H0V2 A0A1Q3F7G3
Pubmed
EMBL
BABH01024416
BABH01024417
NWSH01000528
PCG75876.1
PCG75877.1
KZ149900
+ More
PZC78576.1 ODYU01012516 SOQ58871.1 KQ460398 KPJ15001.1 GAIX01000782 JAA91778.1 RSAL01000003 RVE54876.1 KQ459302 KPJ01802.1 GDQN01011151 JAT79903.1 GDQN01007331 JAT83723.1 GDQN01010989 JAT80065.1 GDQN01009241 JAT81813.1 JTDY01000121 KOB78736.1 GBYB01008177 GBYB01008178 JAG77944.1 JAG77945.1 DS235001 EEB10201.1 NEVH01016952 PNF24896.1 KQ981920 KYN32988.1 KK107570 EZA48836.1 QOIP01000005 RLU22607.1 GL436519 EFN71845.1 KQ978579 KYN30030.1 LBMM01002587 KMQ94611.1 GL448740 EFN83953.1 KQ982612 KYQ53840.1 ADTU01017523 KQ976453 KYM85415.1 GL767674 EFZ13176.1 KQ978255 KYM95882.1 JR036636 JR036637 AEY57280.1 AEY57281.1 JR036639 AEY57282.1 NNAY01002929 OXU20277.1 GBBI01004024 JAC14688.1 GANO01002172 JAB57699.1 GEDC01025493 GEDC01024363 GEDC01024332 JAS11805.1 JAS12935.1 JAS12966.1 PYGN01000420 PSN46691.1 GFTR01005567 JAW10859.1 GEDC01020394 GEDC01000414 JAS16904.1 JAS36884.1 GAKP01020733 JAC38219.1 GAKP01020730 JAC38222.1 GECL01001416 JAP04708.1 GAKP01020731 JAC38221.1 CH479184 EDW36967.1 CH379058 KRT03539.1 EDY69798.1 KQ435916 KOX68750.1 KRT03541.1 KRT03540.1 GAKP01020732 GAKP01020729 JAC38223.1 GECZ01002048 JAS67721.1 GECZ01026504 JAS43265.1 GECZ01027367 JAS42402.1 GECZ01026477 JAS43292.1 GDHF01015599 GDHF01003774 JAI36715.1 JAI48540.1 OUUW01000053 SPP90037.1 GALX01004607 JAB63859.1 GDHF01009433 JAI42881.1 KC763801 AHJ81366.1 GBXI01009495 JAD04797.1 GBXI01007507 JAD06785.1 KA646736 AFP61365.1 GEZM01010784 JAV93836.1 APGK01058894 KB741292 ENN70333.1 KB632276 ERL91173.1 UFQT01000047 SSX18916.1 SSX18915.1 GBHO01005321 JAG38283.1 GBHO01005324 JAG38280.1 GGFK01005142 MBW38463.1 GGFK01005098 MBW38419.1 GGFJ01001391 MBW50532.1 GBHO01018035 GBHO01005326 JAG25569.1 JAG38278.1 GBHO01005323 JAG38281.1 GDIP01112181 JAL91533.1 GGFK01004597 MBW37918.1 GGFK01004512 MBW37833.1 GDIP01227768 JAI95633.1 GDIP01195023 JAJ28379.1 GAMC01018725 GAMC01018724 JAB87831.1 GDIP01195022 JAJ28380.1 GDIP01189173 JAJ34229.1 JXUM01102013 JXUM01102014 JXUM01102015 JXUM01102016 KQ564693 KXJ71890.1 GFDL01011617 JAV23428.1
PZC78576.1 ODYU01012516 SOQ58871.1 KQ460398 KPJ15001.1 GAIX01000782 JAA91778.1 RSAL01000003 RVE54876.1 KQ459302 KPJ01802.1 GDQN01011151 JAT79903.1 GDQN01007331 JAT83723.1 GDQN01010989 JAT80065.1 GDQN01009241 JAT81813.1 JTDY01000121 KOB78736.1 GBYB01008177 GBYB01008178 JAG77944.1 JAG77945.1 DS235001 EEB10201.1 NEVH01016952 PNF24896.1 KQ981920 KYN32988.1 KK107570 EZA48836.1 QOIP01000005 RLU22607.1 GL436519 EFN71845.1 KQ978579 KYN30030.1 LBMM01002587 KMQ94611.1 GL448740 EFN83953.1 KQ982612 KYQ53840.1 ADTU01017523 KQ976453 KYM85415.1 GL767674 EFZ13176.1 KQ978255 KYM95882.1 JR036636 JR036637 AEY57280.1 AEY57281.1 JR036639 AEY57282.1 NNAY01002929 OXU20277.1 GBBI01004024 JAC14688.1 GANO01002172 JAB57699.1 GEDC01025493 GEDC01024363 GEDC01024332 JAS11805.1 JAS12935.1 JAS12966.1 PYGN01000420 PSN46691.1 GFTR01005567 JAW10859.1 GEDC01020394 GEDC01000414 JAS16904.1 JAS36884.1 GAKP01020733 JAC38219.1 GAKP01020730 JAC38222.1 GECL01001416 JAP04708.1 GAKP01020731 JAC38221.1 CH479184 EDW36967.1 CH379058 KRT03539.1 EDY69798.1 KQ435916 KOX68750.1 KRT03541.1 KRT03540.1 GAKP01020732 GAKP01020729 JAC38223.1 GECZ01002048 JAS67721.1 GECZ01026504 JAS43265.1 GECZ01027367 JAS42402.1 GECZ01026477 JAS43292.1 GDHF01015599 GDHF01003774 JAI36715.1 JAI48540.1 OUUW01000053 SPP90037.1 GALX01004607 JAB63859.1 GDHF01009433 JAI42881.1 KC763801 AHJ81366.1 GBXI01009495 JAD04797.1 GBXI01007507 JAD06785.1 KA646736 AFP61365.1 GEZM01010784 JAV93836.1 APGK01058894 KB741292 ENN70333.1 KB632276 ERL91173.1 UFQT01000047 SSX18916.1 SSX18915.1 GBHO01005321 JAG38283.1 GBHO01005324 JAG38280.1 GGFK01005142 MBW38463.1 GGFK01005098 MBW38419.1 GGFJ01001391 MBW50532.1 GBHO01018035 GBHO01005326 JAG25569.1 JAG38278.1 GBHO01005323 JAG38281.1 GDIP01112181 JAL91533.1 GGFK01004597 MBW37918.1 GGFK01004512 MBW37833.1 GDIP01227768 JAI95633.1 GDIP01195023 JAJ28379.1 GAMC01018725 GAMC01018724 JAB87831.1 GDIP01195022 JAJ28380.1 GDIP01189173 JAJ34229.1 JXUM01102013 JXUM01102014 JXUM01102015 JXUM01102016 KQ564693 KXJ71890.1 GFDL01011617 JAV23428.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000283053
UP000053268
UP000037510
+ More
UP000009046 UP000235965 UP000078541 UP000053097 UP000279307 UP000000311 UP000078492 UP000036403 UP000008237 UP000075809 UP000005205 UP000078540 UP000078542 UP000005203 UP000215335 UP000245037 UP000008744 UP000001819 UP000053105 UP000095300 UP000268350 UP000019118 UP000030742 UP000095301 UP000069272 UP000069940 UP000249989
UP000009046 UP000235965 UP000078541 UP000053097 UP000279307 UP000000311 UP000078492 UP000036403 UP000008237 UP000075809 UP000005205 UP000078540 UP000078542 UP000005203 UP000215335 UP000245037 UP000008744 UP000001819 UP000053105 UP000095300 UP000268350 UP000019118 UP000030742 UP000095301 UP000069272 UP000069940 UP000249989
PRIDE
Interpro
SUPFAM
SSF56042
SSF56042
Gene 3D
ProteinModelPortal
H9JBN0
A0A2A4JVR8
A0A2A4JV16
A0A2W1C0T8
A0A2H1X0N8
A0A194RGE1
+ More
S4PNM5 A0A3S2NMN8 A0A194Q8D9 A0A1E1VYT7 A0A1E1W9R8 A0A1E1VZA2 A0A1E1W4A5 A0A0L7LU51 A0A0C9R5V9 E0V9Z5 A0A2J7Q8K9 A0A195EXN5 A0A026VZC9 A0A3L8DQB9 E2A3Z3 A0A151JS49 A0A0J7KWE0 E2BK45 A0A151X0D9 A0A158NJ70 A0A195BL26 E9J1U1 A0A151I9Y5 V9I9X8 A0A088AJM3 V9I9F6 A0A232EPM6 A0A023EZK9 U5EUT9 A0A1B6CHY3 A0A2P8YR13 A0A224XSA3 A0A1B6EG82 A0A034V6V3 A0A034V811 A0A0V0G9V9 A0A034V4P0 B4GK01 A0A0R3NXJ6 B5DHM1 A0A0M8ZQ00 A0A0R3NWU7 A0A0R3NR62 A0A034V9L1 A0A1B6GZ72 A0A1B6EZ88 A0A1I8Q555 A0A1B6EWT1 A0A1I8Q583 A0A1B6EZ74 A0A1I8Q549 A0A0K8WCF1 A0A3B0K757 V5GIH2 A0A0K8VVH9 X2D8A5 A0A0A1X155 A0A0A1X7V9 T1PDA2 A0A1Y1NC82 N6SYN6 U4UFN3 A0A1I8MTF7 A0A1I8MTF6 A0A336LLR6 A0A1I8MTF9 A0A336LMH3 A0A0A9Z4M0 A0A0A9Z8Z9 A0A2M4ACF5 A0A2M4ACC6 A0A2M4BBZ3 A0A0A9Z4L5 A0A182FNC4 A0A0A9Z323 A0A0P5UMR5 A0A2M4ABD1 A0A2M4AAM1 A0A0P4YH45 A0A0P5ASR5 W8B3J4 A0A0P5BUA7 A0A0P5B5V2 A0A182H0V2 A0A1Q3F7G3
S4PNM5 A0A3S2NMN8 A0A194Q8D9 A0A1E1VYT7 A0A1E1W9R8 A0A1E1VZA2 A0A1E1W4A5 A0A0L7LU51 A0A0C9R5V9 E0V9Z5 A0A2J7Q8K9 A0A195EXN5 A0A026VZC9 A0A3L8DQB9 E2A3Z3 A0A151JS49 A0A0J7KWE0 E2BK45 A0A151X0D9 A0A158NJ70 A0A195BL26 E9J1U1 A0A151I9Y5 V9I9X8 A0A088AJM3 V9I9F6 A0A232EPM6 A0A023EZK9 U5EUT9 A0A1B6CHY3 A0A2P8YR13 A0A224XSA3 A0A1B6EG82 A0A034V6V3 A0A034V811 A0A0V0G9V9 A0A034V4P0 B4GK01 A0A0R3NXJ6 B5DHM1 A0A0M8ZQ00 A0A0R3NWU7 A0A0R3NR62 A0A034V9L1 A0A1B6GZ72 A0A1B6EZ88 A0A1I8Q555 A0A1B6EWT1 A0A1I8Q583 A0A1B6EZ74 A0A1I8Q549 A0A0K8WCF1 A0A3B0K757 V5GIH2 A0A0K8VVH9 X2D8A5 A0A0A1X155 A0A0A1X7V9 T1PDA2 A0A1Y1NC82 N6SYN6 U4UFN3 A0A1I8MTF7 A0A1I8MTF6 A0A336LLR6 A0A1I8MTF9 A0A336LMH3 A0A0A9Z4M0 A0A0A9Z8Z9 A0A2M4ACF5 A0A2M4ACC6 A0A2M4BBZ3 A0A0A9Z4L5 A0A182FNC4 A0A0A9Z323 A0A0P5UMR5 A0A2M4ABD1 A0A2M4AAM1 A0A0P4YH45 A0A0P5ASR5 W8B3J4 A0A0P5BUA7 A0A0P5B5V2 A0A182H0V2 A0A1Q3F7G3
Ontologies
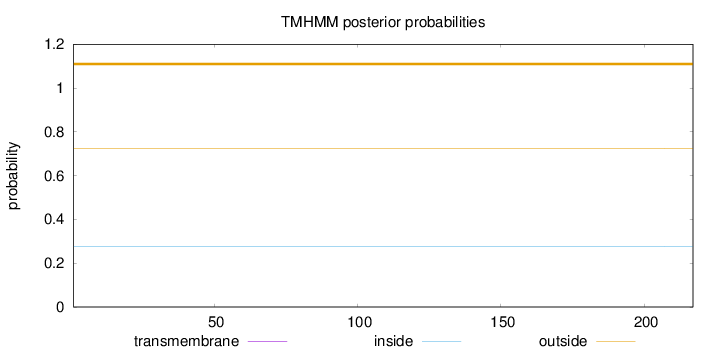
Topology
Length:
217
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.27668
outside
1 - 217
Population Genetic Test Statistics
Pi
260.202075
Theta
203.890966
Tajima's D
1.089153
CLR
0.175762
CSRT
0.687115644217789
Interpretation
Uncertain
