Pre Gene Modal
BGIBMGA008205
Annotation
cationic_peptide_CP8_precursor_[Bombyx_mandarina]
Full name
Fungal protease inhibitor-1
Location in the cell
Extracellular Reliability : 4.529
Sequence
CDS
ATGAAAACGTTGATTTTTATAATGCTGGTGGCATGCGTTGCGTCAGCAGCGTACGGTGCTTTGGTATGTGGTACAGACTACTGTGAAAAAAATCCATGCATACAGCCACCACTTGTTTGCCCTAAGAACACTGAGCATCGCGCCAGACACGCTGGAAAGTGTGCTTGTTGTCCCGCTTGTGTAACTTTACTTGGTGAAGGTGCAACTTGCAAGATTTATTCGAAGGAACTAGGCGAAACCCCCTCCGCTGTGTGTAAGGAGCCTCTCAAATGCATCAAAAGAGTTTGCACTAAGCTTGTGTAG
Protein
MKTLIFIMLVACVASAAYGALVCGTDYCEKNPCIQPPLVCPKNTEHRARHAGKCACCPACVTLLGEGATCKIYSKELGETPSAVCKEPLKCIKRVCTKLV
Summary
Description
Inhibits proteases from the fungi A.oryzae and R.oryzae, trypsin and chymotrypsin. Does not inhibit protease from the bacterium B.licheniformis or papain.
Similarity
Belongs to the membrane-bound acyltransferase family.
Keywords
3D-structure
Direct protein sequencing
Disulfide bond
Protease inhibitor
Serine protease inhibitor
Signal
Feature
chain Fungal protease inhibitor-1
Uniprot
Pubmed
EMBL
BABH01041601
EF126182
ABL76064.1
AY655143
AAU08172.1
AY387408
+ More
AAQ96281.1 GAIX01002767 JAA89793.1 AK402470 BAM19092.1 KR270485 ALH21153.1 KQ460398 KPJ15007.1 AGBW02013372 OWR43513.1 KP713776 AKR15718.1 AM939570 EU375895 CAP74000.1 GDQN01005698 GDQN01001000 JAT85356.1 JAT90054.1 HM448399 ADI87454.1 KQ459302 KPJ01805.1 EF467286 ABO60878.1 AK402024 BAM18646.1 RSAL01000269 RVE43204.1 ODYU01012660 SOQ59089.1 KP792965 AKT09041.1 KC480132 AGR53495.1 PYGN01002990 PSN29770.1 APGK01035488 APGK01035489 APGK01035490 APGK01035491 KB740925 ENN78055.1 HAHE01000092 SNX33522.1 KK852655 KDR19286.1 KB631924 ERL87251.1 CAEY01000613 PYGN01000780 PSN40890.1 PSN40898.1
AAQ96281.1 GAIX01002767 JAA89793.1 AK402470 BAM19092.1 KR270485 ALH21153.1 KQ460398 KPJ15007.1 AGBW02013372 OWR43513.1 KP713776 AKR15718.1 AM939570 EU375895 CAP74000.1 GDQN01005698 GDQN01001000 JAT85356.1 JAT90054.1 HM448399 ADI87454.1 KQ459302 KPJ01805.1 EF467286 ABO60878.1 AK402024 BAM18646.1 RSAL01000269 RVE43204.1 ODYU01012660 SOQ59089.1 KP792965 AKT09041.1 KC480132 AGR53495.1 PYGN01002990 PSN29770.1 APGK01035488 APGK01035489 APGK01035490 APGK01035491 KB740925 ENN78055.1 HAHE01000092 SNX33522.1 KK852655 KDR19286.1 KB631924 ERL87251.1 CAEY01000613 PYGN01000780 PSN40890.1 PSN40898.1
Proteomes
Interpro
ProteinModelPortal
PDB
3BT4
E-value=1.95563e-15,
Score=194
Ontologies
PANTHER
Topology
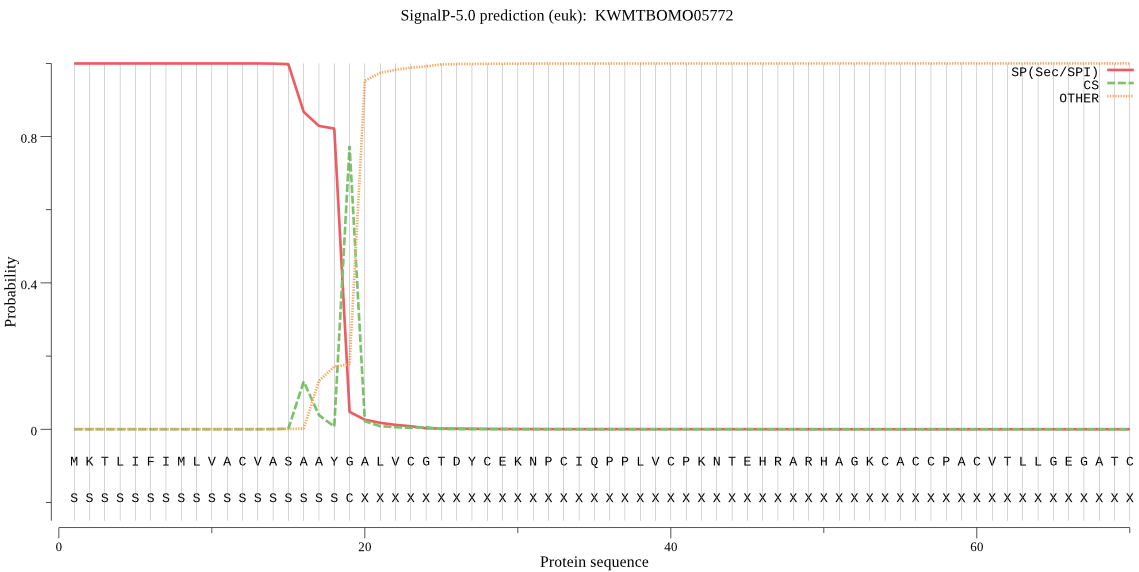
SignalP
Position: 1 - 19,
Likelihood: 0.999741
Length:
100
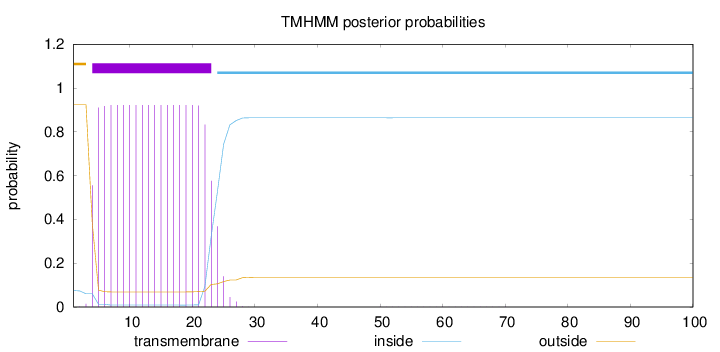
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.23008
Exp number, first 60 AAs:
18.22664
Total prob of N-in:
0.07542
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 23
inside
24 - 100
Population Genetic Test Statistics
Pi
202.089201
Theta
212.930919
Tajima's D
-0.635442
CLR
0.005638
CSRT
0.212089395530223
Interpretation
Uncertain
