Pre Gene Modal
BGIBMGA008210
Annotation
syndecan_binding_protein_[Bombyx_mori]
Full name
Syntenin-1
Alternative Name
Syndecan-binding protein 1
Location in the cell
Nuclear Reliability : 1.785
Sequence
CDS
ATGTCGTTCTATCCGTCTCTGGAGGATATGAAGGTGGATAATATGATGAGGGCTCAAATATCGCAGCATCAAGCACCACCGTCATATGTTGCTCCGCAACTTTGTGCCACGCCTAGTGCCCCTAGTGCTACTCATGTTTACCCAACATTGGGAGAATATATGGGCATGGAGCTGTCCCAGGCTGTAATTGCGCTTAACATGCCCGAATACCAAATACAACAGGTTCAGCCAACAAGTAGTAATGTGGTTGCACCTCTATCATCCCAGTCCCTTTCCCTGCCTAAGGCAACTGTAACTCAGGCTATCAGACAAGTTGTTCTTTGCAAGGACAGAAATGGAAAGTGTGGCTTAAGACTCCATTCAGTTGACAGTGGAGTATTTGTATGCTATGTTGCTGCTAACAGTCCTGGTGCCTTGGCCGGCTTAAGGTTTGGCGATCAGATATTAGAGATCAATAATGTCACTGTTGCTGGAATGACAATGGATAAATGTCATGATATCCTTAAGAAAGCTCCTGCAAATAATATCACCATGGCTGTTCGTGATAGGCCATTCGAGAGAAATGTAACTTTACACAAAGACTCTTTGGGTCATGTGGGATTCCAATTCAAAAATGGAAAAATTATAGCATTGGTTGTTGATTCATCTGCAGCTCGGAATGGCTTATTGACAGACCACCAGATACTGGAAATAAACACAATAAATGTCGTTGGCATGAAAGATAAGGAAATATCTAAAATTATTGATGAAAGTCCTTCTGTTGTAAACATTACAATAATACCTTATTGTATATATGAACGTATGATAAACAAGATGTCATCATCGCTTTTCAAGGAACTGGATCGCACACCAGCAGCTTGA
Protein
MSFYPSLEDMKVDNMMRAQISQHQAPPSYVAPQLCATPSAPSATHVYPTLGEYMGMELSQAVIALNMPEYQIQQVQPTSSNVVAPLSSQSLSLPKATVTQAIRQVVLCKDRNGKCGLRLHSVDSGVFVCYVAANSPGALAGLRFGDQILEINNVTVAGMTMDKCHDILKKAPANNITMAVRDRPFERNVTLHKDSLGHVGFQFKNGKIIALVVDSSAARNGLLTDHQILEINTINVVGMKDKEISKIIDESPSVVNITIIPYCIYERMINKMSSSLFKELDRTPAA
Summary
Description
Multifunctional adapter protein involved in diverse array of functions including trafficking of transmembrane proteins, neuro and immunomodulation, exosome biogenesis, and tumorigenesis. Positively regulates TGFB1-mediated SMAD2/3 activation and TGFB1-induced epithelial-to-mesenchymal transition (EMT) and cell migration in various cell types. May increase TGFB1 signaling by enhancing cell-surface expression of TGFR1 by preventing the interaction between TGFR1 and CAV1 and subsequent CAV1-dependent internalization and degradation of TGFR1. In concert with SDC1/4 and PDCD6IP, regulates exosome biogenesis. Regulates migration, growth, proliferation, and cell cycle progression in a variety of cancer types. In adherens junctions may function to couple syndecans to cytoskeletal proteins or signaling components. Seems to couple transcription factor SOX4 to the IL-5 receptor (IL5RA). May also play a role in vesicular trafficking. Seems to be required for the targeting of TGFA to the cell surface in the early secretory pathway.
Subunit
Monomer and homodimer (PubMed:11152476). Interacts with NFASC and SDCBP2 (PubMed:11152476). Interacts with PTPRJ (PubMed:11434923). Interacts with SDC1, SDC2, SDC3, SDC4, NRXN2, EPHA7, EPHB1, NF2 isoform 1, TGFA and IL5RA (By similarity). Interacts with PDCD6IP. Forms a complex with PDCD6IP and SDC2. Interacts (via C-terminus) with TGFBR1 (By similarity). Binds to FZD7; this interaction is increased by inositol trisphosphate (IP3) (By similarity). Interacts with SMO (By similarity).
Keywords
3D-structure
Acetylation
Cell junction
Cell membrane
Complete proteome
Cytoplasm
Cytoskeleton
Endoplasmic reticulum
Lipid-binding
Membrane
Nucleus
Phosphoprotein
Reference proteome
Repeat
Secreted
Feature
chain Syntenin-1
Uniprot
Q1HQB3
H9JFB3
S4PHU1
A0A2W1BU28
A0A2A4J8Q9
A0A2H1V4T0
+ More
A0A212EPT7 A0A194RC05 I4DJC0 I4DRU3 A0A194QEB4 A0A0L7LM16 V5GR61 A0A1Y1NIR6 E2A3P2 A0A195B096 F4WWC8 A0A158NKJ9 A0A195FC63 A0A151JAZ7 A0A0C9R8J1 A0A026WHA8 A0A151WRJ4 A0A3L8D4U5 A0A0L7R749 D6WZL0 A0A154PF97 A0A1W4X1P6 A0A323TJV5 A0A151IJT2 A0A088ASW2 G3CFQ8 G3CFQ6 A0A0H3WJS6 A0A2A3E9M6 A0A0J7L1E2 G3CFQ7 A0A2J7PQC5 A0A3R7M2V6 A0A2I9LPT4 E9IFD3 E0VM48 A0A1B6DAU4 A0A1B6CEC6 A0A1J1HZE6 A0A1W7R9H5 A0A067R7I7 A0A0P4WQX0 Q17A85 A0A293N5B2 A0A0P5W2M7 B0W419 A0A0P4VTA7 Q1HQS5 C1BQC8 A0A182HDG5 A0A1Q3FCH2 A0A224XSS7 T1JPS3 A0A0N8D8W0 A0A0V0G4C5 T1HMY3 A0A023F2D2 E9G509 A0A182IMS3 A0A023ENS9 A0A069DSB4 A0A0P6H5I2 D3PHC1 T1E1K0 A0A384DD49 A0A0P5ZIX0 A0A2Y9GWX7 A0A1L8FT58 T1ISZ9 A0A091P5K9 A0A3Q7Y560 A0A2Y9KPY0 A0A091NUP6 A0A2M4A464 A0A3Q7X4J0 F1PKR0 A0A3Q7SVB1 A0A384DDU4 G1KE37 A0A1U7TJM1 A0A2Y9H1B8 G3T671 M3X7T2 W5J470 Q9JI92 A0A2U3Y780 A0A0B8RTI2 A0A2Y9KPX8 A0A3R7M2W4 A0A3Q7UJR7 A0A3Q7NY40 A0A0A9YSV5 A0A093PA80 A0A2I0MWZ7
A0A212EPT7 A0A194RC05 I4DJC0 I4DRU3 A0A194QEB4 A0A0L7LM16 V5GR61 A0A1Y1NIR6 E2A3P2 A0A195B096 F4WWC8 A0A158NKJ9 A0A195FC63 A0A151JAZ7 A0A0C9R8J1 A0A026WHA8 A0A151WRJ4 A0A3L8D4U5 A0A0L7R749 D6WZL0 A0A154PF97 A0A1W4X1P6 A0A323TJV5 A0A151IJT2 A0A088ASW2 G3CFQ8 G3CFQ6 A0A0H3WJS6 A0A2A3E9M6 A0A0J7L1E2 G3CFQ7 A0A2J7PQC5 A0A3R7M2V6 A0A2I9LPT4 E9IFD3 E0VM48 A0A1B6DAU4 A0A1B6CEC6 A0A1J1HZE6 A0A1W7R9H5 A0A067R7I7 A0A0P4WQX0 Q17A85 A0A293N5B2 A0A0P5W2M7 B0W419 A0A0P4VTA7 Q1HQS5 C1BQC8 A0A182HDG5 A0A1Q3FCH2 A0A224XSS7 T1JPS3 A0A0N8D8W0 A0A0V0G4C5 T1HMY3 A0A023F2D2 E9G509 A0A182IMS3 A0A023ENS9 A0A069DSB4 A0A0P6H5I2 D3PHC1 T1E1K0 A0A384DD49 A0A0P5ZIX0 A0A2Y9GWX7 A0A1L8FT58 T1ISZ9 A0A091P5K9 A0A3Q7Y560 A0A2Y9KPY0 A0A091NUP6 A0A2M4A464 A0A3Q7X4J0 F1PKR0 A0A3Q7SVB1 A0A384DDU4 G1KE37 A0A1U7TJM1 A0A2Y9H1B8 G3T671 M3X7T2 W5J470 Q9JI92 A0A2U3Y780 A0A0B8RTI2 A0A2Y9KPX8 A0A3R7M2W4 A0A3Q7UJR7 A0A3Q7NY40 A0A0A9YSV5 A0A093PA80 A0A2I0MWZ7
Pubmed
19121390
23622113
28756777
22118469
26354079
22651552
+ More
26227816 28004739 20798317 21719571 21347285 24508170 30249741 18362917 19820115 25731918 29248469 21282665 20566863 24845553 17510324 27129103 17204158 26483478 25474469 21292972 24945155 26334808 24330624 27762356 16341006 24813606 17975172 20920257 23761445 11152476 11179419 15489334 11434923 22673903 25476704 25401762 26823975 23371554
26227816 28004739 20798317 21719571 21347285 24508170 30249741 18362917 19820115 25731918 29248469 21282665 20566863 24845553 17510324 27129103 17204158 26483478 25474469 21292972 24945155 26334808 24330624 27762356 16341006 24813606 17975172 20920257 23761445 11152476 11179419 15489334 11434923 22673903 25476704 25401762 26823975 23371554
EMBL
DQ443139
ABF51228.1
BABH01042044
GAIX01005695
JAA86865.1
KZ149900
+ More
PZC78582.1 NWSH01002617 PCG67840.1 ODYU01000666 SOQ35837.1 AGBW02013372 OWR43510.1 KQ460398 KPJ15009.1 AK401388 BAM18010.1 AK405271 BAM20633.1 KQ459302 KPJ01806.1 JTDY01000682 KOB76241.1 GALX01004389 JAB64077.1 GEZM01003486 JAV96780.1 GL436457 EFN71934.1 KQ976692 KYM77881.1 GL888406 EGI61470.1 ADTU01018869 KQ981693 KYN37792.1 KQ979182 KYN22321.1 GBYB01004410 JAG74177.1 KK107249 EZA54459.1 KQ982805 KYQ50486.1 QOIP01000013 RLU15244.1 KQ414646 KOC66581.1 KQ971372 EFA09687.1 KQ434890 KZC10482.1 MDVM02000127 PYZ99266.1 KQ977294 KYN03894.1 HM210774 AEE65457.1 HM210772 AEE65455.1 KP241935 AKM12661.1 KZ288334 PBC27876.1 LBMM01001444 KMQ96304.1 HM210773 AEE65456.1 NEVH01022639 PNF18537.1 QCYY01002440 ROT70332.1 GFWZ01000400 MBW20390.1 GL762838 EFZ20729.1 DS235289 EEB14454.1 GEDC01014536 JAS22762.1 GEDC01025527 JAS11771.1 CVRI01000037 CRK93479.1 GFAH01000599 JAV47790.1 KK852824 KDR15460.1 GDRN01046530 JAI66775.1 CH477339 EAT43140.1 EAT43141.1 GFWV01022651 MAA47378.1 GDIP01092658 JAM11057.1 DS231834 EDS32612.1 GDKW01000137 JAI56458.1 DQ440369 ABF18402.1 BT076807 ACO11231.1 JXUM01034334 KQ561020 KXJ80005.1 GFDL01009795 JAV25250.1 GFTR01004844 JAW11582.1 CAEY01000429 GDIP01057367 LRGB01000568 JAM46348.1 KZS17869.1 GECL01003212 JAP02912.1 ACPB03015113 GBBI01003548 JAC15164.1 GL732532 EFX85462.1 GAPW01002506 JAC11092.1 GBGD01002268 JAC86621.1 GDIQ01024326 JAN70411.1 BT121027 ADD37957.1 GALA01001594 JAA93258.1 GDIP01042921 JAM60794.1 CM004477 OCT74753.1 JH431451 KK845081 KFP86850.1 KL394081 KFP92659.1 GGFK01002199 MBW35520.1 AAEX03015810 AANG04003518 ADMH02002162 ETN58158.1 AF248548 AJ292243 BC064651 GBSH01002676 JAG66351.1 QCYY01002439 ROT70347.1 GBHO01009436 GDHC01019615 JAG34168.1 JAP99013.1 KL412027 KFW73843.1 AKCR02000001 PKK34206.1
PZC78582.1 NWSH01002617 PCG67840.1 ODYU01000666 SOQ35837.1 AGBW02013372 OWR43510.1 KQ460398 KPJ15009.1 AK401388 BAM18010.1 AK405271 BAM20633.1 KQ459302 KPJ01806.1 JTDY01000682 KOB76241.1 GALX01004389 JAB64077.1 GEZM01003486 JAV96780.1 GL436457 EFN71934.1 KQ976692 KYM77881.1 GL888406 EGI61470.1 ADTU01018869 KQ981693 KYN37792.1 KQ979182 KYN22321.1 GBYB01004410 JAG74177.1 KK107249 EZA54459.1 KQ982805 KYQ50486.1 QOIP01000013 RLU15244.1 KQ414646 KOC66581.1 KQ971372 EFA09687.1 KQ434890 KZC10482.1 MDVM02000127 PYZ99266.1 KQ977294 KYN03894.1 HM210774 AEE65457.1 HM210772 AEE65455.1 KP241935 AKM12661.1 KZ288334 PBC27876.1 LBMM01001444 KMQ96304.1 HM210773 AEE65456.1 NEVH01022639 PNF18537.1 QCYY01002440 ROT70332.1 GFWZ01000400 MBW20390.1 GL762838 EFZ20729.1 DS235289 EEB14454.1 GEDC01014536 JAS22762.1 GEDC01025527 JAS11771.1 CVRI01000037 CRK93479.1 GFAH01000599 JAV47790.1 KK852824 KDR15460.1 GDRN01046530 JAI66775.1 CH477339 EAT43140.1 EAT43141.1 GFWV01022651 MAA47378.1 GDIP01092658 JAM11057.1 DS231834 EDS32612.1 GDKW01000137 JAI56458.1 DQ440369 ABF18402.1 BT076807 ACO11231.1 JXUM01034334 KQ561020 KXJ80005.1 GFDL01009795 JAV25250.1 GFTR01004844 JAW11582.1 CAEY01000429 GDIP01057367 LRGB01000568 JAM46348.1 KZS17869.1 GECL01003212 JAP02912.1 ACPB03015113 GBBI01003548 JAC15164.1 GL732532 EFX85462.1 GAPW01002506 JAC11092.1 GBGD01002268 JAC86621.1 GDIQ01024326 JAN70411.1 BT121027 ADD37957.1 GALA01001594 JAA93258.1 GDIP01042921 JAM60794.1 CM004477 OCT74753.1 JH431451 KK845081 KFP86850.1 KL394081 KFP92659.1 GGFK01002199 MBW35520.1 AAEX03015810 AANG04003518 ADMH02002162 ETN58158.1 AF248548 AJ292243 BC064651 GBSH01002676 JAG66351.1 QCYY01002439 ROT70347.1 GBHO01009436 GDHC01019615 JAG34168.1 JAP99013.1 KL412027 KFW73843.1 AKCR02000001 PKK34206.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000000311 UP000078540 UP000007755 UP000005205 UP000078541 UP000078492 UP000053097 UP000075809 UP000279307 UP000053825 UP000007266 UP000076502 UP000192223 UP000247427 UP000078542 UP000005203 UP000242457 UP000036403 UP000235965 UP000283509 UP000009046 UP000183832 UP000027135 UP000008820 UP000002320 UP000069940 UP000249989 UP000015104 UP000076858 UP000015103 UP000000305 UP000075880 UP000261680 UP000248481 UP000186698 UP000286642 UP000248482 UP000002254 UP000286640 UP000291021 UP000001646 UP000189704 UP000007646 UP000011712 UP000000673 UP000002494 UP000245341 UP000286641 UP000053872
UP000000311 UP000078540 UP000007755 UP000005205 UP000078541 UP000078492 UP000053097 UP000075809 UP000279307 UP000053825 UP000007266 UP000076502 UP000192223 UP000247427 UP000078542 UP000005203 UP000242457 UP000036403 UP000235965 UP000283509 UP000009046 UP000183832 UP000027135 UP000008820 UP000002320 UP000069940 UP000249989 UP000015104 UP000076858 UP000015103 UP000000305 UP000075880 UP000261680 UP000248481 UP000186698 UP000286642 UP000248482 UP000002254 UP000286640 UP000291021 UP000001646 UP000189704 UP000007646 UP000011712 UP000000673 UP000002494 UP000245341 UP000286641 UP000053872
SUPFAM
SSF50156
SSF50156
ProteinModelPortal
Q1HQB3
H9JFB3
S4PHU1
A0A2W1BU28
A0A2A4J8Q9
A0A2H1V4T0
+ More
A0A212EPT7 A0A194RC05 I4DJC0 I4DRU3 A0A194QEB4 A0A0L7LM16 V5GR61 A0A1Y1NIR6 E2A3P2 A0A195B096 F4WWC8 A0A158NKJ9 A0A195FC63 A0A151JAZ7 A0A0C9R8J1 A0A026WHA8 A0A151WRJ4 A0A3L8D4U5 A0A0L7R749 D6WZL0 A0A154PF97 A0A1W4X1P6 A0A323TJV5 A0A151IJT2 A0A088ASW2 G3CFQ8 G3CFQ6 A0A0H3WJS6 A0A2A3E9M6 A0A0J7L1E2 G3CFQ7 A0A2J7PQC5 A0A3R7M2V6 A0A2I9LPT4 E9IFD3 E0VM48 A0A1B6DAU4 A0A1B6CEC6 A0A1J1HZE6 A0A1W7R9H5 A0A067R7I7 A0A0P4WQX0 Q17A85 A0A293N5B2 A0A0P5W2M7 B0W419 A0A0P4VTA7 Q1HQS5 C1BQC8 A0A182HDG5 A0A1Q3FCH2 A0A224XSS7 T1JPS3 A0A0N8D8W0 A0A0V0G4C5 T1HMY3 A0A023F2D2 E9G509 A0A182IMS3 A0A023ENS9 A0A069DSB4 A0A0P6H5I2 D3PHC1 T1E1K0 A0A384DD49 A0A0P5ZIX0 A0A2Y9GWX7 A0A1L8FT58 T1ISZ9 A0A091P5K9 A0A3Q7Y560 A0A2Y9KPY0 A0A091NUP6 A0A2M4A464 A0A3Q7X4J0 F1PKR0 A0A3Q7SVB1 A0A384DDU4 G1KE37 A0A1U7TJM1 A0A2Y9H1B8 G3T671 M3X7T2 W5J470 Q9JI92 A0A2U3Y780 A0A0B8RTI2 A0A2Y9KPX8 A0A3R7M2W4 A0A3Q7UJR7 A0A3Q7NY40 A0A0A9YSV5 A0A093PA80 A0A2I0MWZ7
A0A212EPT7 A0A194RC05 I4DJC0 I4DRU3 A0A194QEB4 A0A0L7LM16 V5GR61 A0A1Y1NIR6 E2A3P2 A0A195B096 F4WWC8 A0A158NKJ9 A0A195FC63 A0A151JAZ7 A0A0C9R8J1 A0A026WHA8 A0A151WRJ4 A0A3L8D4U5 A0A0L7R749 D6WZL0 A0A154PF97 A0A1W4X1P6 A0A323TJV5 A0A151IJT2 A0A088ASW2 G3CFQ8 G3CFQ6 A0A0H3WJS6 A0A2A3E9M6 A0A0J7L1E2 G3CFQ7 A0A2J7PQC5 A0A3R7M2V6 A0A2I9LPT4 E9IFD3 E0VM48 A0A1B6DAU4 A0A1B6CEC6 A0A1J1HZE6 A0A1W7R9H5 A0A067R7I7 A0A0P4WQX0 Q17A85 A0A293N5B2 A0A0P5W2M7 B0W419 A0A0P4VTA7 Q1HQS5 C1BQC8 A0A182HDG5 A0A1Q3FCH2 A0A224XSS7 T1JPS3 A0A0N8D8W0 A0A0V0G4C5 T1HMY3 A0A023F2D2 E9G509 A0A182IMS3 A0A023ENS9 A0A069DSB4 A0A0P6H5I2 D3PHC1 T1E1K0 A0A384DD49 A0A0P5ZIX0 A0A2Y9GWX7 A0A1L8FT58 T1ISZ9 A0A091P5K9 A0A3Q7Y560 A0A2Y9KPY0 A0A091NUP6 A0A2M4A464 A0A3Q7X4J0 F1PKR0 A0A3Q7SVB1 A0A384DDU4 G1KE37 A0A1U7TJM1 A0A2Y9H1B8 G3T671 M3X7T2 W5J470 Q9JI92 A0A2U3Y780 A0A0B8RTI2 A0A2Y9KPX8 A0A3R7M2W4 A0A3Q7UJR7 A0A3Q7NY40 A0A0A9YSV5 A0A093PA80 A0A2I0MWZ7
PDB
5G1E
E-value=1.44286e-59,
Score=580
Ontologies
GO
GO:0035556
GO:0030036
GO:0007268
GO:0005886
GO:0005737
GO:0010862
GO:0030511
GO:0045545
GO:0005137
GO:0031965
GO:0005829
GO:0008284
GO:0005654
GO:0030307
GO:0007265
GO:0005895
GO:0030335
GO:0016021
GO:0042470
GO:0010718
GO:0046875
GO:0046982
GO:0008289
GO:0042803
GO:0070062
GO:0050839
GO:0008022
GO:0042043
GO:0019838
GO:0099054
GO:0005789
GO:0045121
GO:0007346
GO:0005856
GO:0032435
GO:0005925
GO:0002091
GO:0005515
GO:0006468
GO:0005524
GO:0009190
GO:0016849
GO:0004568
GO:0006032
GO:0006541
GO:0016812
GO:0016597
PANTHER
Topology
Subcellular location
Cell junction
Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Focal adhesion Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Adherens junction Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Cell membrane Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Endoplasmic reticulum membrane Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Nucleus Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Melanosome Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Cytoplasm Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Cytosol Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Cytoskeleton Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Secreted Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Exosome Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Membrane raft Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Focal adhesion Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Adherens junction Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Cell membrane Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Endoplasmic reticulum membrane Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Nucleus Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Melanosome Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Cytoplasm Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Cytosol Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Cytoskeleton Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Secreted Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Exosome Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
Membrane raft Mainly membrane-associated. Localized to adherens junctions, focal adhesions and endoplasmic reticulum. Colocalized with actin stress fibers. Also found in the nucleus. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2) (By similarity). With evidence from 5 publications.
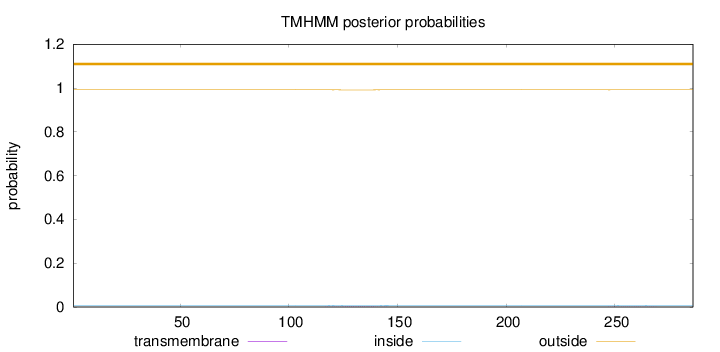
Length:
286
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0474000000000001
Exp number, first 60 AAs:
0.0013
Total prob of N-in:
0.00797
outside
1 - 286
Population Genetic Test Statistics
Pi
215.488297
Theta
190.171481
Tajima's D
0.568805
CLR
0
CSRT
0.534623268836558
Interpretation
Uncertain
