Gene
KWMTBOMO05762
Pre Gene Modal
BGIBMGA006801
Annotation
PREDICTED:_guanylate_cyclase_32E-like_[Bombyx_mori]
Full name
Guanylate cyclase
+ More
Receptor-type guanylate cyclase Gyc76C
Receptor-type guanylate cyclase Gyc76C
Alternative Name
DrGC-1
Location in the cell
PlasmaMembrane Reliability : 2.638
Sequence
CDS
ATGTGGCACGGTGCGCGGGTGTTGGTGTTCGCGGCGCTGGGCTTACTGGCTGGGCCTTCGCTTGCCCACAAGAAGTTCAATATCACCATTGGCTACTTACCGGCTCTTACCGGTGACTTGCGAGACAGACAAGGACTCACAGTCTCCGGGGCACTGTCGTTGGCACTTGAAGAGCTGAACAATGGAGTCGATCTGCTGCCGGATGTGGAGATTCACATGCAGTGGAACAACACGAAATATGACATGGTAACTTCCACCAGGCTCATCACCGAAATGAGTCGTGCAGGTGTAGTAGCCTTTTTCGGACCCGAAGGTCGCTGTGAAACTGAAGCCATAATTGCGCAATCCAGAAACTTGCCTATGATCTCTTACAAATGCTCAGACTACCAAGTATCTAGGCTACCAAACTTTGCTCGATTAGAGCCTCCGGGCGTTCAGGCGACAAAAGCGTTAATATCGTTGCTACGTTACTATAGATGGAACAAATTCTCTATATTGTATGAAGAATCTTGGATGACGGTAGCCCTGACGTTGGCGAACTATGCCAAGGATTTTAACATGACTGTAAACCATCAGAGTCCCGTCATAAATACTTTTAAATGTTGCGTAGAGAAAATGGCTTGCTGTGCACCCGGTTTCTGGTATCAGTTCATTCATGATACAAAAAACAAAACAAGAATTTACGTTTTCCTTGGGTCGACTTCGGGACTAATAGAGATGATGACCGCCATGGAGTCGCTGCAGTTATTCGCGAAGGGCGAGTACATGGTTATATTCGTGGACATGATGACGTACTCTCTTAAAGACTCTGCCAAGTATTTGTTGAAAATCGAAGATCGTTTATCGTCGACATTAATATGTCCGAACACGCCTGAATTTAAAAAGAGAGCCCAGTCGTTACTGGTGATAGCTTCGTCGGAACCATCCCCGGAATATGAAGAGTTTACGGCTAAAGTCAGGAAATACAATTCTCTAGAACCATTCAATTTTCCCGATGTCTTTGGCAGCAAGTACATTAAGTTTGTATCCATTTACGCTGCGTACTTGTATGATTCGGTGAAGCTCTATGCTAAAGCTCTAGATGAAATAATCAAGGAGGAAACTCAAAAGGAGCCCTTGACGCAGGCCAAGCTAACGAATATTGTCACTAACGGAACAGCCATTGTGGCGAAGATGATTAAAAGTTCGCCATACAAAAGTGTGTCCGGTTTGACGATACGTTTAGATGATCGTGGCGATTCCGAGGGAAACTTCTCCTTGCTCGCTTTCAAGCCGATCAACAACAAGATTGATAGAAACTGCAGTTACAGTATGATACCGGTTGCTCATTTCCAGCAAAACAGTAAAGAGTTTCCTGATTACAAACTGAATTCTCGTATACCGATCGATTGGCCTGACAGCGTGAAACCGGAGGACGAACCGTCTTGCGGTTTCAACAACGAGAAATGTCCAAGGCACAACTCGCTGATAACGTTGGCGTTACCGTTAGCGTTGACGGGAACGTTGGCGGTGACTTTGTTTTGTGTGTTATTCGTCACGGTCAGCAAATACCGAAAATGGAAGATCAAACAAGAAATAGAAGGTTTTCTTTGGGAAATAGACGCACAGGAAGTTACCGGATATTCCGACAACGGCCTCGTGTGCTCGCCCAGTAAGTTATGTCTAGCGAGCGGTATATCATACGAAATGTGCCCACAAAAATTCACCACGACGGCGCAATACAGAGGGAATTTAGTCAGGATAAAAACATTGAAGTTTTCTAAAAAGAAGGATATATCTCGGGACATAATGAAAGAGATGCGGCTGCTGCGCGAGCTGCGTCACGACAATCTCAACTCGTTCATCGGCGCCGTCGTGGAGCCGATGCGCGTGCTCATCCTCACCGACTACTGTGCCAAGGGGAGCTTGTATGATATAATAGAAAACGAAGATATCAAATTGGATCAGATGTTAATATCTTTGCTTATTCACGATCTCATTAAGGGTATGATCTTCATTCATTCATCGCCTTTAATATTTCACGGAAATCTCAAATCCTCGAATTGTGTAGTTACATCAAGATGGATGTTACAAGTAGCTGATTTTGATCTTCGTGAGCTGAAATATTGCGCTGAAAACAAGTTTATTGGAGAACATCAGTATTATAGAGGTTTGCTTTGGAAATCACCAGAGTTGTTGAGAGGATTAAACAACCCGAATGGCATCGTGGGCGGAACGCAAAAAGGTGACGTCTATGCGTTTGGGATTATTCTATACGAAGTGATAGCGAGAAAAGGCCCTTTCGGAAATGTCGCAATGGAGCCCAAAGATATAATAGACCGCTTGAAGCGGCCGAAGTTGGACGGCGAAGAGTTGTTTCGTCCGGACACGAGCAGCATCGACGGCGGCATCGACGACTACATCGTGTCGTGCATGAAGGACTGCTGGGCAGAAGATCCCAACACAAGACCTGACTTCCCTACGATTAGAACCAGACTAAAGAAGATGAAATCGGTACAGTAA
Protein
MWHGARVLVFAALGLLAGPSLAHKKFNITIGYLPALTGDLRDRQGLTVSGALSLALEELNNGVDLLPDVEIHMQWNNTKYDMVTSTRLITEMSRAGVVAFFGPEGRCETEAIIAQSRNLPMISYKCSDYQVSRLPNFARLEPPGVQATKALISLLRYYRWNKFSILYEESWMTVALTLANYAKDFNMTVNHQSPVINTFKCCVEKMACCAPGFWYQFIHDTKNKTRIYVFLGSTSGLIEMMTAMESLQLFAKGEYMVIFVDMMTYSLKDSAKYLLKIEDRLSSTLICPNTPEFKKRAQSLLVIASSEPSPEYEEFTAKVRKYNSLEPFNFPDVFGSKYIKFVSIYAAYLYDSVKLYAKALDEIIKEETQKEPLTQAKLTNIVTNGTAIVAKMIKSSPYKSVSGLTIRLDDRGDSEGNFSLLAFKPINNKIDRNCSYSMIPVAHFQQNSKEFPDYKLNSRIPIDWPDSVKPEDEPSCGFNNEKCPRHNSLITLALPLALTGTLAVTLFCVLFVTVSKYRKWKIKQEIEGFLWEIDAQEVTGYSDNGLVCSPSKLCLASGISYEMCPQKFTTTAQYRGNLVRIKTLKFSKKKDISRDIMKEMRLLRELRHDNLNSFIGAVVEPMRVLILTDYCAKGSLYDIIENEDIKLDQMLISLLIHDLIKGMIFIHSSPLIFHGNLKSSNCVVTSRWMLQVADFDLRELKYCAENKFIGEHQYYRGLLWKSPELLRGLNNPNGIVGGTQKGDVYAFGIILYEVIARKGPFGNVAMEPKDIIDRLKRPKLDGEELFRPDTSSIDGGIDDYIVSCMKDCWAEDPNTRPDFPTIRTRLKKMKSVQ
Summary
Description
Guanylate cyclase involved in the production of the second messenger cGMP (PubMed:24284209). Acts as a receptor for the NPLP1-4 peptide and modulates the innate immune IMD pathway in response to salt stress by inducing nuclear translocation of NF-kappa-B protein Rel which leads to increased expression of the antimicrobial peptide diptericin (PubMed:21893139). Plays a role in Sema-1a-mediated axon repulsion which is required for the correct establishment of neuromuscular connectivity (PubMed:15282266, PubMed:24284209). Required in developing embryonic somatic muscle for correct patterning of ventral and lateral muscles and for localization of integrin beta-ps at developing dorsal muscle myotendinous junctions (PubMed:23213443). Required for invagination, migration and lumen shape of the embryonic salivary gland by regulating the localization of the integrin-binding protein rhea/Talin to the visceral mesoderm surrounding the gland and maintaining the laminin matrix (PubMed:23862019). Required in the developing wing to regulate extracellular matrix (ECM) organization by activating the cGMP-dependent protein kinase For which represses the activity of matrix metalloproteases such as Mmp2 and decreases ECM matrix reorganization (PubMed:26440503).
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Subunit
Interacts with the semaphorin 1A receptor PlexA; PlexA enhances Gyc76C catalytic activity. Interacts with the PDZ domain-containing protein kermit; kermit increases cell surface expression of Gyc76C.
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Keywords
ATP-binding
Cell membrane
cGMP biosynthesis
Complete proteome
Glycoprotein
GTP-binding
Lyase
Magnesium
Membrane
Metal-binding
Nucleotide-binding
Receptor
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain Guanylate cyclase
Uniprot
H9JBA6
H9JBJ6
A0A3S2LS11
A0A2H1WKG2
A0A194RAY4
A0A2A4J2G4
+ More
A0A2W1BY57 A0A194Q8F6 A0A212F645 S4P787 A0A1L8DJG1 A0A1L8DJE6 A0A1L8DJP0 A0A1L8DJP4 A0A1L8DJR8 A0A1L8DJF6 D6X177 E0VCX6 A0A0L0C8M8 T1PCH4 W8AIX8 A0A1I8PNZ6 A0A1W4XT11 A0A1Y1M579 A0A034V957 A0A1J1IPW9 A0A1A9ZDW0 A0A1A9UKP9 Q16YZ6 A0A182HEV4 A0A1A9XJW6 A0A1B0C6N8 A0A0A1X4C8 A0A0A1WZK5 A0A084WB37 A0A1B0GF71 B4N3M7 A0A1B0CUZ5 A0A182WD90 A0A1A9W5X4 Q7PZZ3 A0A182PGB7 A0A182HPR9 A0A0M4EG81 A0A182X009 A0A182KT21 A0A182J455 B5DRL7 B4LGE0 A0A3B0J659 A0A182QXR8 B4KY29 A0A2M3YZT9 A4V243 Q7JQ32 B3M6J3 B4QQX8 B3NII0 A0A182FW72 A0A2M4A487 A0A2M4A468 B4PFY8 A0A2M4A4N6 B4IWW8 A0A1W4UDX7 K7J3D3 A0A2M4CIM0 A0A2M4CKB0 A0A2M4CLW1 A0A182RXD1 A0A0L7RJ80 A0A3L8DCZ8 A0A151X5N2 A0A088AVQ7 A0A158NJ88 A0A151JZJ7 A0A336LXX4 A0A182MXC5 A0A067QVM8 B4IIP9 A0A1B6EEF4 A0A1B6MKA2 A0A1B6F1B7 U4TZM5 N6UEX4 A0A2H8TK31 J9JNT2 A0A224XIA1 A0A2S2R4A9 A0A023F3B8 A0A3Q0J1D4 A0A2P8YZ06 A0A182YQ82 A0A0M8ZPG3 B0WSE8 E9J8Y6 A0A195BKT2 A0A151IK46 T1IAR7 A0A026WMG4
A0A2W1BY57 A0A194Q8F6 A0A212F645 S4P787 A0A1L8DJG1 A0A1L8DJE6 A0A1L8DJP0 A0A1L8DJP4 A0A1L8DJR8 A0A1L8DJF6 D6X177 E0VCX6 A0A0L0C8M8 T1PCH4 W8AIX8 A0A1I8PNZ6 A0A1W4XT11 A0A1Y1M579 A0A034V957 A0A1J1IPW9 A0A1A9ZDW0 A0A1A9UKP9 Q16YZ6 A0A182HEV4 A0A1A9XJW6 A0A1B0C6N8 A0A0A1X4C8 A0A0A1WZK5 A0A084WB37 A0A1B0GF71 B4N3M7 A0A1B0CUZ5 A0A182WD90 A0A1A9W5X4 Q7PZZ3 A0A182PGB7 A0A182HPR9 A0A0M4EG81 A0A182X009 A0A182KT21 A0A182J455 B5DRL7 B4LGE0 A0A3B0J659 A0A182QXR8 B4KY29 A0A2M3YZT9 A4V243 Q7JQ32 B3M6J3 B4QQX8 B3NII0 A0A182FW72 A0A2M4A487 A0A2M4A468 B4PFY8 A0A2M4A4N6 B4IWW8 A0A1W4UDX7 K7J3D3 A0A2M4CIM0 A0A2M4CKB0 A0A2M4CLW1 A0A182RXD1 A0A0L7RJ80 A0A3L8DCZ8 A0A151X5N2 A0A088AVQ7 A0A158NJ88 A0A151JZJ7 A0A336LXX4 A0A182MXC5 A0A067QVM8 B4IIP9 A0A1B6EEF4 A0A1B6MKA2 A0A1B6F1B7 U4TZM5 N6UEX4 A0A2H8TK31 J9JNT2 A0A224XIA1 A0A2S2R4A9 A0A023F3B8 A0A3Q0J1D4 A0A2P8YZ06 A0A182YQ82 A0A0M8ZPG3 B0WSE8 E9J8Y6 A0A195BKT2 A0A151IK46 T1IAR7 A0A026WMG4
EC Number
4.6.1.2
Pubmed
19121390
26354079
28756777
22118469
23622113
18362917
+ More
19820115 20566863 26108605 25315136 24495485 28004739 25348373 17510324 26483478 25830018 24438588 17994087 12364791 20966253 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7706258 7759483 15282266 23213443 21893139 23862019 24284209 26440503 22936249 17550304 20075255 30249741 21347285 24845553 23537049 25474469 29403074 25244985 21282665 24508170
19820115 20566863 26108605 25315136 24495485 28004739 25348373 17510324 26483478 25830018 24438588 17994087 12364791 20966253 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7706258 7759483 15282266 23213443 21893139 23862019 24284209 26440503 22936249 17550304 20075255 30249741 21347285 24845553 23537049 25474469 29403074 25244985 21282665 24508170
EMBL
BABH01030967
BABH01030968
BABH01030969
BABH01030970
BABH01030971
BABH01030972
+ More
RSAL01000275 RVE43120.1 ODYU01009272 SOQ53570.1 KQ460398 KPJ15013.1 NWSH01003611 PCG66059.1 KZ149900 PZC78585.1 KQ459302 KPJ01808.1 AGBW02010077 OWR49210.1 GAIX01004724 JAA87836.1 GFDF01007498 JAV06586.1 GFDF01007496 JAV06588.1 GFDF01007499 JAV06585.1 GFDF01007497 JAV06587.1 GFDF01007500 JAV06584.1 GFDF01007501 JAV06583.1 KQ971372 EFA10586.2 DS235068 EEB11232.1 JRES01000760 KNC28587.1 KA645835 AFP60464.1 GAMC01020881 GAMC01020880 GAMC01020878 JAB85677.1 GEZM01043506 JAV79046.1 GAKP01019888 GAKP01019887 JAC39064.1 CVRI01000055 CRL01142.1 CH477506 EAT39830.1 JXUM01036120 JXUM01036121 JXUM01036122 JXUM01036123 JXUM01036124 JXUM01036125 JXUM01036126 JXUM01036127 KQ561084 KXJ79765.1 JXJN01026843 GBXI01015827 GBXI01008764 GBXI01005765 JAC98464.1 JAD05528.1 JAD08527.1 GBXI01010499 JAD03793.1 ATLV01022307 ATLV01022308 KE525331 KFB47431.1 CCAG010012768 CH964095 EDW79232.1 AJWK01029883 AJWK01029884 AJWK01029885 AJWK01029886 AJWK01029887 AJWK01029888 AJWK01029889 AAAB01008986 EAA00176.4 APCN01003270 CP012525 ALC45096.1 CH379070 EDY74228.1 CH940647 EDW70469.1 OUUW01000002 SPP77407.1 AXCN02001683 CH933809 EDW18731.1 GGFM01001015 MBW21766.1 AE014296 AAF49117.1 AAV50029.1 L35598 U23485 BT011062 AAA74408.1 AAA85858.1 AAR31133.1 CH902618 EDV40842.1 CM000363 CM002912 EDX11116.1 KMZ00596.1 CH954178 EDV52405.1 GGFK01002303 MBW35624.1 GGFK01002283 MBW35604.1 CM000159 EDW95285.1 GGFK01002453 MBW35774.1 CH916366 EDV97369.1 GGFL01001032 MBW65210.1 GGFL01001598 MBW65776.1 GGFL01001700 MBW65878.1 KQ414582 KOC70843.1 QOIP01000010 RLU18186.1 KQ982498 KYQ55696.1 ADTU01017581 ADTU01017582 ADTU01017583 ADTU01017584 ADTU01017585 ADTU01017586 ADTU01017587 KQ981427 KYN41775.1 UFQT01000216 SSX21841.1 KK853196 KDR09972.1 CH480844 EDW49820.1 GEDC01007580 GEDC01000969 JAS29718.1 JAS36329.1 GEBQ01016200 GEBQ01014835 GEBQ01003621 JAT23777.1 JAT25142.1 JAT36356.1 GECZ01025815 GECZ01004426 JAS43954.1 JAS65343.1 KB631805 ERL86282.1 APGK01029504 APGK01029505 APGK01029506 KB740735 ENN79171.1 GFXV01002712 MBW14517.1 ABLF02031542 ABLF02031545 ABLF02043285 GFTR01008655 JAW07771.1 GGMS01015560 MBY84763.1 GBBI01002797 JAC15915.1 PYGN01000278 PSN49479.1 KQ435922 KOX68415.1 DS232069 EDS33793.1 GL769162 EFZ10727.1 KQ976453 KYM85379.1 KQ977255 KYN04643.1 ACPB03019543 KK107167 EZA56309.1
RSAL01000275 RVE43120.1 ODYU01009272 SOQ53570.1 KQ460398 KPJ15013.1 NWSH01003611 PCG66059.1 KZ149900 PZC78585.1 KQ459302 KPJ01808.1 AGBW02010077 OWR49210.1 GAIX01004724 JAA87836.1 GFDF01007498 JAV06586.1 GFDF01007496 JAV06588.1 GFDF01007499 JAV06585.1 GFDF01007497 JAV06587.1 GFDF01007500 JAV06584.1 GFDF01007501 JAV06583.1 KQ971372 EFA10586.2 DS235068 EEB11232.1 JRES01000760 KNC28587.1 KA645835 AFP60464.1 GAMC01020881 GAMC01020880 GAMC01020878 JAB85677.1 GEZM01043506 JAV79046.1 GAKP01019888 GAKP01019887 JAC39064.1 CVRI01000055 CRL01142.1 CH477506 EAT39830.1 JXUM01036120 JXUM01036121 JXUM01036122 JXUM01036123 JXUM01036124 JXUM01036125 JXUM01036126 JXUM01036127 KQ561084 KXJ79765.1 JXJN01026843 GBXI01015827 GBXI01008764 GBXI01005765 JAC98464.1 JAD05528.1 JAD08527.1 GBXI01010499 JAD03793.1 ATLV01022307 ATLV01022308 KE525331 KFB47431.1 CCAG010012768 CH964095 EDW79232.1 AJWK01029883 AJWK01029884 AJWK01029885 AJWK01029886 AJWK01029887 AJWK01029888 AJWK01029889 AAAB01008986 EAA00176.4 APCN01003270 CP012525 ALC45096.1 CH379070 EDY74228.1 CH940647 EDW70469.1 OUUW01000002 SPP77407.1 AXCN02001683 CH933809 EDW18731.1 GGFM01001015 MBW21766.1 AE014296 AAF49117.1 AAV50029.1 L35598 U23485 BT011062 AAA74408.1 AAA85858.1 AAR31133.1 CH902618 EDV40842.1 CM000363 CM002912 EDX11116.1 KMZ00596.1 CH954178 EDV52405.1 GGFK01002303 MBW35624.1 GGFK01002283 MBW35604.1 CM000159 EDW95285.1 GGFK01002453 MBW35774.1 CH916366 EDV97369.1 GGFL01001032 MBW65210.1 GGFL01001598 MBW65776.1 GGFL01001700 MBW65878.1 KQ414582 KOC70843.1 QOIP01000010 RLU18186.1 KQ982498 KYQ55696.1 ADTU01017581 ADTU01017582 ADTU01017583 ADTU01017584 ADTU01017585 ADTU01017586 ADTU01017587 KQ981427 KYN41775.1 UFQT01000216 SSX21841.1 KK853196 KDR09972.1 CH480844 EDW49820.1 GEDC01007580 GEDC01000969 JAS29718.1 JAS36329.1 GEBQ01016200 GEBQ01014835 GEBQ01003621 JAT23777.1 JAT25142.1 JAT36356.1 GECZ01025815 GECZ01004426 JAS43954.1 JAS65343.1 KB631805 ERL86282.1 APGK01029504 APGK01029505 APGK01029506 KB740735 ENN79171.1 GFXV01002712 MBW14517.1 ABLF02031542 ABLF02031545 ABLF02043285 GFTR01008655 JAW07771.1 GGMS01015560 MBY84763.1 GBBI01002797 JAC15915.1 PYGN01000278 PSN49479.1 KQ435922 KOX68415.1 DS232069 EDS33793.1 GL769162 EFZ10727.1 KQ976453 KYM85379.1 KQ977255 KYN04643.1 ACPB03019543 KK107167 EZA56309.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000218220
UP000053268
UP000007151
+ More
UP000007266 UP000009046 UP000037069 UP000095301 UP000095300 UP000192223 UP000183832 UP000092445 UP000078200 UP000008820 UP000069940 UP000249989 UP000092443 UP000092460 UP000030765 UP000092444 UP000007798 UP000092461 UP000075920 UP000091820 UP000007062 UP000075885 UP000075840 UP000092553 UP000076407 UP000075882 UP000075880 UP000001819 UP000008792 UP000268350 UP000075886 UP000009192 UP000000803 UP000007801 UP000000304 UP000008711 UP000069272 UP000002282 UP000001070 UP000192221 UP000002358 UP000075900 UP000053825 UP000279307 UP000075809 UP000005203 UP000005205 UP000078541 UP000075884 UP000027135 UP000001292 UP000030742 UP000019118 UP000007819 UP000079169 UP000245037 UP000076408 UP000053105 UP000002320 UP000078540 UP000078542 UP000015103 UP000053097
UP000007266 UP000009046 UP000037069 UP000095301 UP000095300 UP000192223 UP000183832 UP000092445 UP000078200 UP000008820 UP000069940 UP000249989 UP000092443 UP000092460 UP000030765 UP000092444 UP000007798 UP000092461 UP000075920 UP000091820 UP000007062 UP000075885 UP000075840 UP000092553 UP000076407 UP000075882 UP000075880 UP000001819 UP000008792 UP000268350 UP000075886 UP000009192 UP000000803 UP000007801 UP000000304 UP000008711 UP000069272 UP000002282 UP000001070 UP000192221 UP000002358 UP000075900 UP000053825 UP000279307 UP000075809 UP000005203 UP000005205 UP000078541 UP000075884 UP000027135 UP000001292 UP000030742 UP000019118 UP000007819 UP000079169 UP000245037 UP000076408 UP000053105 UP000002320 UP000078540 UP000078542 UP000015103 UP000053097
Interpro
Gene 3D
ProteinModelPortal
H9JBA6
H9JBJ6
A0A3S2LS11
A0A2H1WKG2
A0A194RAY4
A0A2A4J2G4
+ More
A0A2W1BY57 A0A194Q8F6 A0A212F645 S4P787 A0A1L8DJG1 A0A1L8DJE6 A0A1L8DJP0 A0A1L8DJP4 A0A1L8DJR8 A0A1L8DJF6 D6X177 E0VCX6 A0A0L0C8M8 T1PCH4 W8AIX8 A0A1I8PNZ6 A0A1W4XT11 A0A1Y1M579 A0A034V957 A0A1J1IPW9 A0A1A9ZDW0 A0A1A9UKP9 Q16YZ6 A0A182HEV4 A0A1A9XJW6 A0A1B0C6N8 A0A0A1X4C8 A0A0A1WZK5 A0A084WB37 A0A1B0GF71 B4N3M7 A0A1B0CUZ5 A0A182WD90 A0A1A9W5X4 Q7PZZ3 A0A182PGB7 A0A182HPR9 A0A0M4EG81 A0A182X009 A0A182KT21 A0A182J455 B5DRL7 B4LGE0 A0A3B0J659 A0A182QXR8 B4KY29 A0A2M3YZT9 A4V243 Q7JQ32 B3M6J3 B4QQX8 B3NII0 A0A182FW72 A0A2M4A487 A0A2M4A468 B4PFY8 A0A2M4A4N6 B4IWW8 A0A1W4UDX7 K7J3D3 A0A2M4CIM0 A0A2M4CKB0 A0A2M4CLW1 A0A182RXD1 A0A0L7RJ80 A0A3L8DCZ8 A0A151X5N2 A0A088AVQ7 A0A158NJ88 A0A151JZJ7 A0A336LXX4 A0A182MXC5 A0A067QVM8 B4IIP9 A0A1B6EEF4 A0A1B6MKA2 A0A1B6F1B7 U4TZM5 N6UEX4 A0A2H8TK31 J9JNT2 A0A224XIA1 A0A2S2R4A9 A0A023F3B8 A0A3Q0J1D4 A0A2P8YZ06 A0A182YQ82 A0A0M8ZPG3 B0WSE8 E9J8Y6 A0A195BKT2 A0A151IK46 T1IAR7 A0A026WMG4
A0A2W1BY57 A0A194Q8F6 A0A212F645 S4P787 A0A1L8DJG1 A0A1L8DJE6 A0A1L8DJP0 A0A1L8DJP4 A0A1L8DJR8 A0A1L8DJF6 D6X177 E0VCX6 A0A0L0C8M8 T1PCH4 W8AIX8 A0A1I8PNZ6 A0A1W4XT11 A0A1Y1M579 A0A034V957 A0A1J1IPW9 A0A1A9ZDW0 A0A1A9UKP9 Q16YZ6 A0A182HEV4 A0A1A9XJW6 A0A1B0C6N8 A0A0A1X4C8 A0A0A1WZK5 A0A084WB37 A0A1B0GF71 B4N3M7 A0A1B0CUZ5 A0A182WD90 A0A1A9W5X4 Q7PZZ3 A0A182PGB7 A0A182HPR9 A0A0M4EG81 A0A182X009 A0A182KT21 A0A182J455 B5DRL7 B4LGE0 A0A3B0J659 A0A182QXR8 B4KY29 A0A2M3YZT9 A4V243 Q7JQ32 B3M6J3 B4QQX8 B3NII0 A0A182FW72 A0A2M4A487 A0A2M4A468 B4PFY8 A0A2M4A4N6 B4IWW8 A0A1W4UDX7 K7J3D3 A0A2M4CIM0 A0A2M4CKB0 A0A2M4CLW1 A0A182RXD1 A0A0L7RJ80 A0A3L8DCZ8 A0A151X5N2 A0A088AVQ7 A0A158NJ88 A0A151JZJ7 A0A336LXX4 A0A182MXC5 A0A067QVM8 B4IIP9 A0A1B6EEF4 A0A1B6MKA2 A0A1B6F1B7 U4TZM5 N6UEX4 A0A2H8TK31 J9JNT2 A0A224XIA1 A0A2S2R4A9 A0A023F3B8 A0A3Q0J1D4 A0A2P8YZ06 A0A182YQ82 A0A0M8ZPG3 B0WSE8 E9J8Y6 A0A195BKT2 A0A151IK46 T1IAR7 A0A026WMG4
PDB
3P86
E-value=1.03098e-25,
Score=293
Ontologies
GO
GO:0004383
GO:0004672
GO:0016021
GO:0005524
GO:0035556
GO:0000166
GO:0006182
GO:0007165
GO:0007168
GO:0001653
GO:0005886
GO:0019934
GO:0038023
GO:0005887
GO:0030215
GO:0030510
GO:0009651
GO:0005525
GO:0007411
GO:0007428
GO:0008045
GO:0016199
GO:0046872
GO:0007526
GO:0030198
GO:0007435
GO:0006468
GO:0019752
GO:0030170
GO:0004568
GO:0006032
GO:0006541
GO:0016812
GO:0016597
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 22,
Likelihood: 0.983461
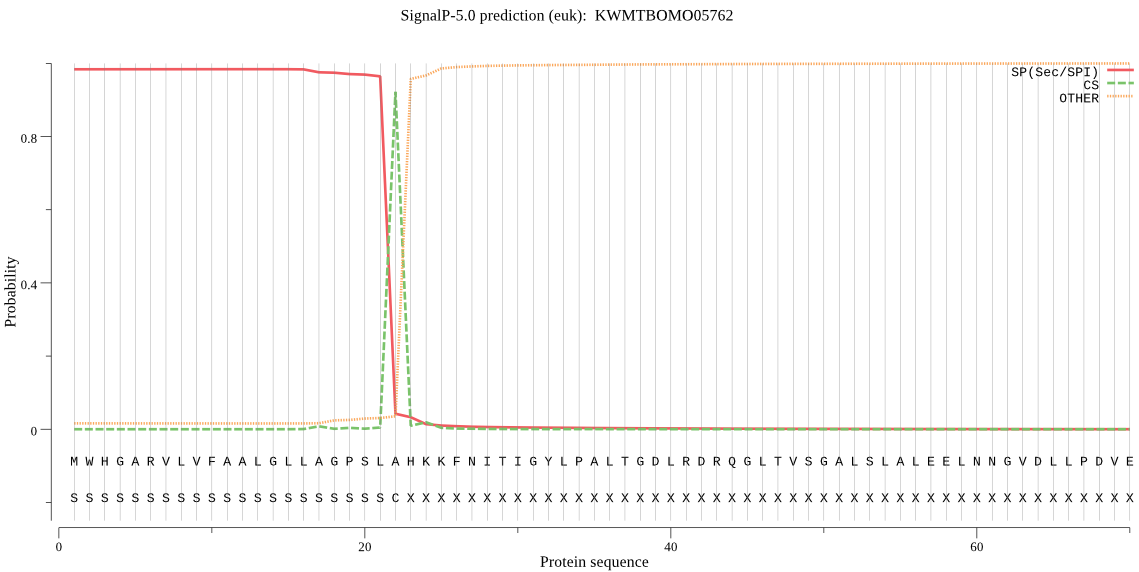
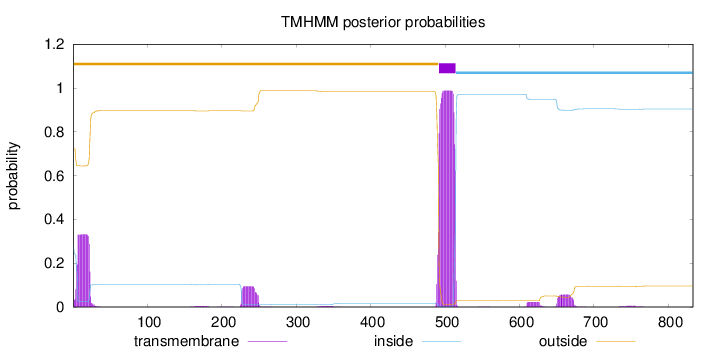
Length:
833
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
32.50022
Exp number, first 60 AAs:
6.07606
Total prob of N-in:
0.27728
outside
1 - 491
TMhelix
492 - 514
inside
515 - 833
Population Genetic Test Statistics
Pi
195.171459
Theta
150.870816
Tajima's D
1.156291
CLR
0.589927
CSRT
0.702364881755912
Interpretation
Uncertain
