Gene
KWMTBOMO05761
Pre Gene Modal
BGIBMGA006889
Annotation
PREDICTED:_T-cell_immunomodulatory_protein_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 3.787
Sequence
CDS
ATGGTGTACACAATTTTTATTTTGTTATTTAATGTAATTTTGTCGACAAGCTCTTTTGTAAGTGATATTACTAGCAGTTCGTTTGGAAAATATTCCGAAGGAATTATAGCTGCATTCGGTGATTTTAATTCTGACGAATTAACTGACGCCTTCGTTATTAAAAATAGTTCAACTGTCAACGTTTTTCTTGCATATGACAAAGAGCCTTTTCTAAGGCCATCCGAATTTGCATGCAACTTTTCTGATATTGTTATCACTAGTATTGTACCAGGAGATTACGATGGAGATGCTTACATGGATATACTATTGACCACTCAAAATGTAAACAATACTTCAAAATTTCACAATGTGAGTATACTTTGGGGAGGTATGCCCACATTAAATTGTTCTGATGCTCTTCAAACGAAAGCAATAACATTTGAACAGCCATTAGTCATGGATTACAATAGAGACATGATCTTAGATTTGTTTGGTATCAATGTTGAGGGCAAACGAGTGTTTTGGATTTTTGACCAGTCACGTAACAAACCCAAAGAAAAACCAATGGGAGATGAAAAATTATACAAAGAAATCAAACAACCTCACTCGCATTCTTTTCTGGACGTAAATGATGATGATGCAGCAGATCTCTTGATAACTACCACATCAGAAGTTGAAGTGTGGCTGAATGAAGATACAAATGGGTTTAAGTATGACTGTAGTATTGAATTTTTGATTGGCCATGCTGCTATTTATGGCCAGGCATTGTTTCTAGATGTTTCTTTATCTGGAGAATTTTTTTTGGCTATACCTGTATGTTATGACATTGATTGTTTCAACAGCACTATTTTAATATATGACAATATTGAGTGGCATGATCTCCAAGTGGACTTTAATGATGGCAAAGGAACGTTATGGAGATTTACTCCTCCAAGAGATGAAATTTACTTAGACACCATTACAATGCGTAGTGGTGATTACAATATGGATGGGTACCCTGATATTTTAATGGTACTAAGTCCGACCCACAGTAATGACACTAGAGTGTTCCTACTTCATAATATACCATGCAATTTACCAGGATGTAAATTCGTCAGAACATTTCAAATACACTGGGATATGTTTGATTCATTTGGTAATGATATTGTAATGGCCACTTTTTACGATTTTTATATGGATGGAGTTTTAGATATAATTTATGTTAAGAAAAATTATACATCTCAAACACACACAATGCATTCATTTAGAAATGAGTTAGAATATGATACCAACTTTATCAAGGTTATTGTGGTCACTGGCTTAACTAATGCTATAGTGCCAATCTTAAACACTACTTTATACAAGAGAAAGGCTACTTTTGGTACTAATTTACCAGGACCGAAAATTGGATACAATACTTGGTCCCAAGAAGGAACTTATAGAACAGGTGTTTGTGCACAGTTACCGCAGTCTGCATATTTTGCTTTACAACTTCCTTATTCCATATTTGGTCTTGACAGAACACCAAACTTTGTGGATACACTAAATGTTGGGCTATCTGGCTATTCCAAAAGTTGGACTCAAATTATTCCAAATTCTCAAATAGTTGTAATACCTGTGCCACCTTATGATCCATCTCAATGGAAAGCACAACTATTTGTTACACCTAGCAAAGTGATTCTCAAAAGTGTTTTTGTACTAACTGCAATTATAGTTATCATAACAGGGTGTGTACTATATTTGCATTGGAAAGAAAGAAATGATAGACAAGATATCATCGAGATTGATGATCAGACCTATGTAAAGATATGA
Protein
MVYTIFILLFNVILSTSSFVSDITSSSFGKYSEGIIAAFGDFNSDELTDAFVIKNSSTVNVFLAYDKEPFLRPSEFACNFSDIVITSIVPGDYDGDAYMDILLTTQNVNNTSKFHNVSILWGGMPTLNCSDALQTKAITFEQPLVMDYNRDMILDLFGINVEGKRVFWIFDQSRNKPKEKPMGDEKLYKEIKQPHSHSFLDVNDDDAADLLITTTSEVEVWLNEDTNGFKYDCSIEFLIGHAAIYGQALFLDVSLSGEFFLAIPVCYDIDCFNSTILIYDNIEWHDLQVDFNDGKGTLWRFTPPRDEIYLDTITMRSGDYNMDGYPDILMVLSPTHSNDTRVFLLHNIPCNLPGCKFVRTFQIHWDMFDSFGNDIVMATFYDFYMDGVLDIIYVKKNYTSQTHTMHSFRNELEYDTNFIKVIVVTGLTNAIVPILNTTLYKRKATFGTNLPGPKIGYNTWSQEGTYRTGVCAQLPQSAYFALQLPYSIFGLDRTPNFVDTLNVGLSGYSKSWTQIIPNSQIVVIPVPPYDPSQWKAQLFVTPSKVILKSVFVLTAIIVIITGCVLYLHWKERNDRQDIIEIDDQTYVKI
Summary
Uniprot
A0A2W1BZB2
A0A2A4JPJ2
A0A194Q9Z3
A0A1E1W1I4
A0A194RCK5
A0A0L7LDD1
+ More
A0A3S2LQC9 H9JBJ4 A0A2H1WL29 A0A139WBG8 A0A0L7QVJ5 E2BU62 A0A0M9A8A0 A0A195D2Z2 A0A2A3EKW4 V9IKM1 A0A087ZWI4 A0A1Y1L1K4 A0A0C9RAE6 A0A1W4WFN0 F4W6H9 E1ZVK4 A0A1A9YG34 E9IHE7 A0A195E416 A0A1B0AR17 A0A1A9V4D5 A0A1A9Z8Z6 A0A1B0FQ46 V5G4D7 D3TMT0 A0A026WJY1 A0A232FFS4 A0A151X244 A0A195FI10 A0A0J7L2H4 K7J0W2 A0A195AVX3 A0A158P286 A0A0A1XD56 A0A034WA38 A0A0K8WJ90 W8ARX9 A0A0L0CDV3 T1P8Z9 A0A1I8M5B7 A0A1I8NPN0 A0A224XLM7 A0A1L8E2V0 U4UQU6 A0A1B0CY08 A0A1B6HT44 A0A1B6IZN2 B0WFT7 A0A084W9A1 A0A1Q3FHF8 A0A2R7WVE1 A0A1B6MFK1 A0A182N2Z4 A0A182QTB4 A0A182JGP7 A0A182GSE5 Q9VUQ7 B3NI39 B4KUJ6 A0A182W223 A0A0M4F0M2 B4HIG3 B4PCH4 A0A182UXI6 A0A182L5A5 A0A182XJN2 Q7PZK9 A0A182HHP7 B4QL35 Q176J4 A0A182LYJ2 A0A2M4BH68 A0A2M4BH76 A0A182NZX8 A0A182YBM5 A0A182RKF5 A0A182KA97 U5EUY6 T1GPQ4 A0A182TAT1 A0A1W4VA72 A0A2H8TVC5 J9JL54 A0A2M3Z577 A0A2J7QHE6 B4NLX1 A0A2M3ZH70 A0A2J7QHF2 A0A2J7QHH4 B4IWV0 A0A2M4AD93 Q2M1C8
A0A3S2LQC9 H9JBJ4 A0A2H1WL29 A0A139WBG8 A0A0L7QVJ5 E2BU62 A0A0M9A8A0 A0A195D2Z2 A0A2A3EKW4 V9IKM1 A0A087ZWI4 A0A1Y1L1K4 A0A0C9RAE6 A0A1W4WFN0 F4W6H9 E1ZVK4 A0A1A9YG34 E9IHE7 A0A195E416 A0A1B0AR17 A0A1A9V4D5 A0A1A9Z8Z6 A0A1B0FQ46 V5G4D7 D3TMT0 A0A026WJY1 A0A232FFS4 A0A151X244 A0A195FI10 A0A0J7L2H4 K7J0W2 A0A195AVX3 A0A158P286 A0A0A1XD56 A0A034WA38 A0A0K8WJ90 W8ARX9 A0A0L0CDV3 T1P8Z9 A0A1I8M5B7 A0A1I8NPN0 A0A224XLM7 A0A1L8E2V0 U4UQU6 A0A1B0CY08 A0A1B6HT44 A0A1B6IZN2 B0WFT7 A0A084W9A1 A0A1Q3FHF8 A0A2R7WVE1 A0A1B6MFK1 A0A182N2Z4 A0A182QTB4 A0A182JGP7 A0A182GSE5 Q9VUQ7 B3NI39 B4KUJ6 A0A182W223 A0A0M4F0M2 B4HIG3 B4PCH4 A0A182UXI6 A0A182L5A5 A0A182XJN2 Q7PZK9 A0A182HHP7 B4QL35 Q176J4 A0A182LYJ2 A0A2M4BH68 A0A2M4BH76 A0A182NZX8 A0A182YBM5 A0A182RKF5 A0A182KA97 U5EUY6 T1GPQ4 A0A182TAT1 A0A1W4VA72 A0A2H8TVC5 J9JL54 A0A2M3Z577 A0A2J7QHE6 B4NLX1 A0A2M3ZH70 A0A2J7QHF2 A0A2J7QHH4 B4IWV0 A0A2M4AD93 Q2M1C8
Pubmed
28756777
26354079
26227816
19121390
18362917
19820115
+ More
20798317 28004739 21719571 21282665 20353571 24508170 30249741 28648823 20075255 21347285 25830018 25348373 24495485 26108605 25315136 23537049 24438588 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 20966253 12364791 14747013 17210077 22936249 17510324 25244985 15632085
20798317 28004739 21719571 21282665 20353571 24508170 30249741 28648823 20075255 21347285 25830018 25348373 24495485 26108605 25315136 23537049 24438588 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 20966253 12364791 14747013 17210077 22936249 17510324 25244985 15632085
EMBL
KZ149900
PZC78587.1
NWSH01000887
PCG73676.1
KQ459302
KPJ01810.1
+ More
GDQN01010265 JAT80789.1 KQ460398 KPJ15015.1 JTDY01001572 KOB73487.1 RSAL01000493 RVE41543.1 BABH01030967 ODYU01009364 SOQ53738.1 KQ971372 KYB25191.1 KQ414727 KOC62579.1 GL450581 EFN80779.1 KQ435713 KOX79270.1 KQ976885 KYN07280.1 KZ288215 PBC32453.1 JR050275 AEY61217.1 GEZM01069063 JAV66668.1 GBYB01013424 GBYB01014624 JAG83191.1 JAG84391.1 GL887707 EGI70314.1 GL434548 EFN74773.1 GL763245 EFZ19994.1 KQ979657 KYN19925.1 JXJN01002206 CCAG010006155 GALX01003566 JAB64900.1 EZ422732 ADD19008.1 KK107207 QOIP01000002 EZA55419.1 RLU26137.1 NNAY01000332 OXU29157.1 KQ982580 KYQ54482.1 KQ981523 KYN40330.1 LBMM01001134 KMQ96848.1 KQ976731 KYM76180.1 ADTU01007024 GBXI01005416 JAD08876.1 GAKP01007388 JAC51564.1 GDHF01001180 JAI51134.1 GAMC01014960 JAB91595.1 JRES01000502 KNC30593.1 KA645172 AFP59801.1 GFTR01007373 JAW09053.1 GFDF01001014 JAV13070.1 KB632328 ERL92531.1 AJWK01035238 GECU01029876 JAS77830.1 GECU01015396 JAS92310.1 DS231920 EDS26431.1 ATLV01021659 KE525321 KFB46795.1 GFDL01008049 JAV26996.1 KK855429 PTY22535.1 GEBQ01005274 JAT34703.1 AXCN02001144 JXUM01084370 KQ563433 KXJ73841.1 AE014296 AY075477 AAF49618.1 AAL68289.1 CH954178 EDV52125.1 CH933809 EDW18224.1 CP012525 ALC44712.1 CH480815 EDW41593.1 CM000159 EDW94884.1 AAAB01008986 EAA00374.3 APCN01002432 CM000363 CM002912 EDX10560.1 KMY99772.1 CH477386 EAT42089.1 AXCM01000173 GGFJ01003268 MBW52409.1 GGFJ01003269 MBW52410.1 GANO01002114 JAB57757.1 CAQQ02174633 GFXV01006399 MBW18204.1 ABLF02021664 GGFM01002915 MBW23666.1 NEVH01013976 PNF28015.1 CH964278 EDW85423.1 GGFM01007059 MBW27810.1 PNF28014.1 PNF28016.1 CH916366 EDV97351.1 GGFK01005435 MBW38756.1 CH379069 EAL30646.3
GDQN01010265 JAT80789.1 KQ460398 KPJ15015.1 JTDY01001572 KOB73487.1 RSAL01000493 RVE41543.1 BABH01030967 ODYU01009364 SOQ53738.1 KQ971372 KYB25191.1 KQ414727 KOC62579.1 GL450581 EFN80779.1 KQ435713 KOX79270.1 KQ976885 KYN07280.1 KZ288215 PBC32453.1 JR050275 AEY61217.1 GEZM01069063 JAV66668.1 GBYB01013424 GBYB01014624 JAG83191.1 JAG84391.1 GL887707 EGI70314.1 GL434548 EFN74773.1 GL763245 EFZ19994.1 KQ979657 KYN19925.1 JXJN01002206 CCAG010006155 GALX01003566 JAB64900.1 EZ422732 ADD19008.1 KK107207 QOIP01000002 EZA55419.1 RLU26137.1 NNAY01000332 OXU29157.1 KQ982580 KYQ54482.1 KQ981523 KYN40330.1 LBMM01001134 KMQ96848.1 KQ976731 KYM76180.1 ADTU01007024 GBXI01005416 JAD08876.1 GAKP01007388 JAC51564.1 GDHF01001180 JAI51134.1 GAMC01014960 JAB91595.1 JRES01000502 KNC30593.1 KA645172 AFP59801.1 GFTR01007373 JAW09053.1 GFDF01001014 JAV13070.1 KB632328 ERL92531.1 AJWK01035238 GECU01029876 JAS77830.1 GECU01015396 JAS92310.1 DS231920 EDS26431.1 ATLV01021659 KE525321 KFB46795.1 GFDL01008049 JAV26996.1 KK855429 PTY22535.1 GEBQ01005274 JAT34703.1 AXCN02001144 JXUM01084370 KQ563433 KXJ73841.1 AE014296 AY075477 AAF49618.1 AAL68289.1 CH954178 EDV52125.1 CH933809 EDW18224.1 CP012525 ALC44712.1 CH480815 EDW41593.1 CM000159 EDW94884.1 AAAB01008986 EAA00374.3 APCN01002432 CM000363 CM002912 EDX10560.1 KMY99772.1 CH477386 EAT42089.1 AXCM01000173 GGFJ01003268 MBW52409.1 GGFJ01003269 MBW52410.1 GANO01002114 JAB57757.1 CAQQ02174633 GFXV01006399 MBW18204.1 ABLF02021664 GGFM01002915 MBW23666.1 NEVH01013976 PNF28015.1 CH964278 EDW85423.1 GGFM01007059 MBW27810.1 PNF28014.1 PNF28016.1 CH916366 EDV97351.1 GGFK01005435 MBW38756.1 CH379069 EAL30646.3
Proteomes
UP000218220
UP000053268
UP000053240
UP000037510
UP000283053
UP000005204
+ More
UP000007266 UP000053825 UP000008237 UP000053105 UP000078542 UP000242457 UP000005203 UP000192223 UP000007755 UP000000311 UP000092443 UP000078492 UP000092460 UP000078200 UP000092445 UP000092444 UP000053097 UP000279307 UP000215335 UP000075809 UP000078541 UP000036403 UP000002358 UP000078540 UP000005205 UP000037069 UP000095301 UP000095300 UP000030742 UP000092461 UP000002320 UP000030765 UP000075884 UP000075886 UP000075880 UP000069940 UP000249989 UP000000803 UP000008711 UP000009192 UP000075920 UP000092553 UP000001292 UP000002282 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000000304 UP000008820 UP000075883 UP000075885 UP000076408 UP000075900 UP000075881 UP000015102 UP000075901 UP000192221 UP000007819 UP000235965 UP000007798 UP000001070 UP000001819
UP000007266 UP000053825 UP000008237 UP000053105 UP000078542 UP000242457 UP000005203 UP000192223 UP000007755 UP000000311 UP000092443 UP000078492 UP000092460 UP000078200 UP000092445 UP000092444 UP000053097 UP000279307 UP000215335 UP000075809 UP000078541 UP000036403 UP000002358 UP000078540 UP000005205 UP000037069 UP000095301 UP000095300 UP000030742 UP000092461 UP000002320 UP000030765 UP000075884 UP000075886 UP000075880 UP000069940 UP000249989 UP000000803 UP000008711 UP000009192 UP000075920 UP000092553 UP000001292 UP000002282 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000000304 UP000008820 UP000075883 UP000075885 UP000076408 UP000075900 UP000075881 UP000015102 UP000075901 UP000192221 UP000007819 UP000235965 UP000007798 UP000001070 UP000001819
Interpro
SUPFAM
SSF82704
SSF82704
Gene 3D
ProteinModelPortal
A0A2W1BZB2
A0A2A4JPJ2
A0A194Q9Z3
A0A1E1W1I4
A0A194RCK5
A0A0L7LDD1
+ More
A0A3S2LQC9 H9JBJ4 A0A2H1WL29 A0A139WBG8 A0A0L7QVJ5 E2BU62 A0A0M9A8A0 A0A195D2Z2 A0A2A3EKW4 V9IKM1 A0A087ZWI4 A0A1Y1L1K4 A0A0C9RAE6 A0A1W4WFN0 F4W6H9 E1ZVK4 A0A1A9YG34 E9IHE7 A0A195E416 A0A1B0AR17 A0A1A9V4D5 A0A1A9Z8Z6 A0A1B0FQ46 V5G4D7 D3TMT0 A0A026WJY1 A0A232FFS4 A0A151X244 A0A195FI10 A0A0J7L2H4 K7J0W2 A0A195AVX3 A0A158P286 A0A0A1XD56 A0A034WA38 A0A0K8WJ90 W8ARX9 A0A0L0CDV3 T1P8Z9 A0A1I8M5B7 A0A1I8NPN0 A0A224XLM7 A0A1L8E2V0 U4UQU6 A0A1B0CY08 A0A1B6HT44 A0A1B6IZN2 B0WFT7 A0A084W9A1 A0A1Q3FHF8 A0A2R7WVE1 A0A1B6MFK1 A0A182N2Z4 A0A182QTB4 A0A182JGP7 A0A182GSE5 Q9VUQ7 B3NI39 B4KUJ6 A0A182W223 A0A0M4F0M2 B4HIG3 B4PCH4 A0A182UXI6 A0A182L5A5 A0A182XJN2 Q7PZK9 A0A182HHP7 B4QL35 Q176J4 A0A182LYJ2 A0A2M4BH68 A0A2M4BH76 A0A182NZX8 A0A182YBM5 A0A182RKF5 A0A182KA97 U5EUY6 T1GPQ4 A0A182TAT1 A0A1W4VA72 A0A2H8TVC5 J9JL54 A0A2M3Z577 A0A2J7QHE6 B4NLX1 A0A2M3ZH70 A0A2J7QHF2 A0A2J7QHH4 B4IWV0 A0A2M4AD93 Q2M1C8
A0A3S2LQC9 H9JBJ4 A0A2H1WL29 A0A139WBG8 A0A0L7QVJ5 E2BU62 A0A0M9A8A0 A0A195D2Z2 A0A2A3EKW4 V9IKM1 A0A087ZWI4 A0A1Y1L1K4 A0A0C9RAE6 A0A1W4WFN0 F4W6H9 E1ZVK4 A0A1A9YG34 E9IHE7 A0A195E416 A0A1B0AR17 A0A1A9V4D5 A0A1A9Z8Z6 A0A1B0FQ46 V5G4D7 D3TMT0 A0A026WJY1 A0A232FFS4 A0A151X244 A0A195FI10 A0A0J7L2H4 K7J0W2 A0A195AVX3 A0A158P286 A0A0A1XD56 A0A034WA38 A0A0K8WJ90 W8ARX9 A0A0L0CDV3 T1P8Z9 A0A1I8M5B7 A0A1I8NPN0 A0A224XLM7 A0A1L8E2V0 U4UQU6 A0A1B0CY08 A0A1B6HT44 A0A1B6IZN2 B0WFT7 A0A084W9A1 A0A1Q3FHF8 A0A2R7WVE1 A0A1B6MFK1 A0A182N2Z4 A0A182QTB4 A0A182JGP7 A0A182GSE5 Q9VUQ7 B3NI39 B4KUJ6 A0A182W223 A0A0M4F0M2 B4HIG3 B4PCH4 A0A182UXI6 A0A182L5A5 A0A182XJN2 Q7PZK9 A0A182HHP7 B4QL35 Q176J4 A0A182LYJ2 A0A2M4BH68 A0A2M4BH76 A0A182NZX8 A0A182YBM5 A0A182RKF5 A0A182KA97 U5EUY6 T1GPQ4 A0A182TAT1 A0A1W4VA72 A0A2H8TVC5 J9JL54 A0A2M3Z577 A0A2J7QHE6 B4NLX1 A0A2M3ZH70 A0A2J7QHF2 A0A2J7QHH4 B4IWV0 A0A2M4AD93 Q2M1C8
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.925114
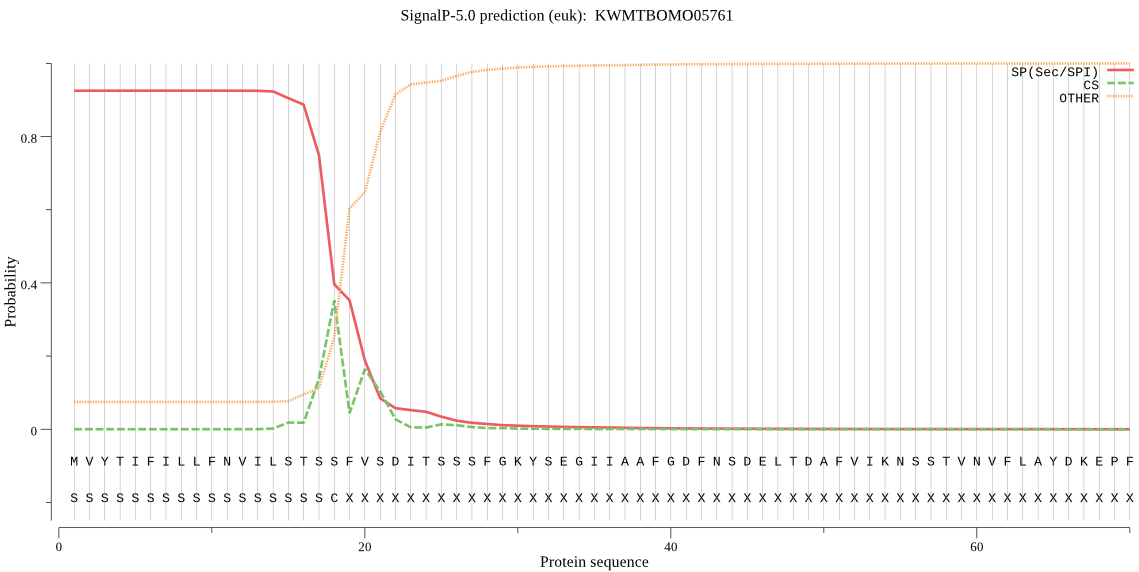
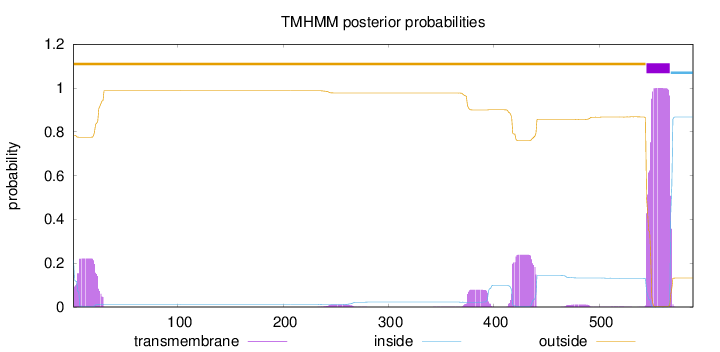
Length:
589
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
33.74513
Exp number, first 60 AAs:
4.63811999999999
Total prob of N-in:
0.21612
outside
1 - 544
TMhelix
545 - 567
inside
568 - 589
Population Genetic Test Statistics
Pi
271.13937
Theta
210.04673
Tajima's D
0.719156
CLR
0.062359
CSRT
0.577471126443678
Interpretation
Uncertain
