Pre Gene Modal
BGIBMGA006802
Annotation
preimplantation_protein_[Bombyx_mori]
Full name
MOB kinase activator-like 4
Alternative Name
Mob as tumor suppressor protein 4
Mps one binder kinase activator-like 4
Mps one binder kinase activator-like 4
Location in the cell
Nuclear Reliability : 2.489
Sequence
CDS
ATGACCGACGGAGTTCAAATACTCCGACGCAATCGACCCGGCACCAAAGCCAAGGACTTCTGTCGCTGGCCTGATGAACCCTTTGAAGAAATGGATAGCACATTGGCTGTTCAACAATTTATTCAACAAACTATTCGACGTGACCCTTCGAATCTCGAAGCTATTTTGAAGATGCCAGAGGTGTTGGATGAAGGTGTTTGGAAGTATGAACATCTAAGACAATTTTGTATGGAGCTCAATGGATTGGCTGTGCGCCTACAAAATGAATGCAAACCTGAAACATGTACACAAATGACTGCAACAGAACAGTGGATATTTCTGTGTGCAGCACATAAAACTCCTAAAGAATGTCCAGCCATTGATTACACACGGCACACTCTAGATGGAGCAGCTTGCTTGTTGAATAGCAATAAATACTTTCCAAGCAGGGTGAACATCAAGGATTCATCAATTGCCAAGTTGGGATCTGTTTGTAGAAGGGTTTACAGGATCTTCTCACATGCATACTTTCATCATCGTTCAATTTATGATGCATTTGAAAGAGAAACTCATTTATGCAAGAGATTCACATATTTTGTAACAAAATACCCAATAATACCTCAAGAAATTCTAATTTTACCTAAACTTGATGATGAAGTACCAGCTGGTGAATCAGAAGCTTGA
Protein
MTDGVQILRRNRPGTKAKDFCRWPDEPFEEMDSTLAVQQFIQQTIRRDPSNLEAILKMPEVLDEGVWKYEHLRQFCMELNGLAVRLQNECKPETCTQMTATEQWIFLCAAHKTPKECPAIDYTRHTLDGAACLLNSNKYFPSRVNIKDSSIAKLGSVCRRVYRIFSHAYFHHRSIYDAFERETHLCKRFTYFVTKYPIIPQEILILPKLDDEVPAGESEA
Summary
Similarity
Belongs to the MOB1/phocein family.
Keywords
Complete proteome
Metal-binding
Reference proteome
Zinc
Feature
chain MOB kinase activator-like 4
Uniprot
Q1HQ19
A0A2H1WL05
A0A194RGF7
A0A194QEB9
A0A1E1WM92
A0A2W1BU66
+ More
A0A2A4JNP9 S4P5J4 A0A0L7LDL2 A0A212F646 D6X397 A0A0T6AV83 A0A1W4XIX0 A0A1A9W102 B4LK09 B3MIP6 A0A0M5IXV4 A0A0C9QK69 B4P101 A0A1I8MQE3 B4NMH0 B4KNI4 B4J6J5 Q292Z2 B4GC78 A0A158NCJ4 A0A1W4V1Z3 A0A0J9R7W7 B4HQA7 B3NB34 Q7K0E3 A0A1I8P7Q1 A0A1B0FC86 A0A1A9YC02 A0A1A9UU23 A0A1B0AKE4 A0A3B0J442 E2BHY1 A0A3L8DHM0 A0A1W7R880 A0A0L0C2V7 B4QDB6 A0A0K8TPQ0 A0A087TML9 W8BEA4 A0A1W4VEA4 A0A0R1DQY9 A0A0Q9WFL3 A0A0J9TUE8 A0A0B4K6X9 A0A0K8RM50 B7PUT2 A0A0Q9XEW9 K7JC07 A0A088AVR4 Q17F92 A0A0K8WAB8 A0A034WPW0 A0A1B6CA79 A0A0P4VQ69 A0A023GGH9 A0A1E1XRB6 A0A154PN19 A0A0A9YPS0 J3JWS5 A0A182KC29 A0A1Q3F580 A0A224XWX8 A0A195F8P7 A0A1Q3F524 B0WT47 T1IBN4 W5JNT7 A0A2R7VW68 A0A2M3Z9Z1 A0A2M4ATB5 A0A182FG47 A0A2M4BYY2 A0A023EK78 A0A023F4F2 A0A0N0BC34 A0A182QEA4 A0A224YJH1 L7M7P3 A0A069DR44 A0A0V0GCC5 A0A182IPU3 A0A161MHQ6 A0A131Y097 A0A2M4ATU6 A0A182M5Q7 A0A182VIC8 A0A182NKM1 A0A182TD06 A0A182WJK4 A0A182KQW3 A0A182WZ28 Q7PF81 A0A182RU03 A0A182I2X8
A0A2A4JNP9 S4P5J4 A0A0L7LDL2 A0A212F646 D6X397 A0A0T6AV83 A0A1W4XIX0 A0A1A9W102 B4LK09 B3MIP6 A0A0M5IXV4 A0A0C9QK69 B4P101 A0A1I8MQE3 B4NMH0 B4KNI4 B4J6J5 Q292Z2 B4GC78 A0A158NCJ4 A0A1W4V1Z3 A0A0J9R7W7 B4HQA7 B3NB34 Q7K0E3 A0A1I8P7Q1 A0A1B0FC86 A0A1A9YC02 A0A1A9UU23 A0A1B0AKE4 A0A3B0J442 E2BHY1 A0A3L8DHM0 A0A1W7R880 A0A0L0C2V7 B4QDB6 A0A0K8TPQ0 A0A087TML9 W8BEA4 A0A1W4VEA4 A0A0R1DQY9 A0A0Q9WFL3 A0A0J9TUE8 A0A0B4K6X9 A0A0K8RM50 B7PUT2 A0A0Q9XEW9 K7JC07 A0A088AVR4 Q17F92 A0A0K8WAB8 A0A034WPW0 A0A1B6CA79 A0A0P4VQ69 A0A023GGH9 A0A1E1XRB6 A0A154PN19 A0A0A9YPS0 J3JWS5 A0A182KC29 A0A1Q3F580 A0A224XWX8 A0A195F8P7 A0A1Q3F524 B0WT47 T1IBN4 W5JNT7 A0A2R7VW68 A0A2M3Z9Z1 A0A2M4ATB5 A0A182FG47 A0A2M4BYY2 A0A023EK78 A0A023F4F2 A0A0N0BC34 A0A182QEA4 A0A224YJH1 L7M7P3 A0A069DR44 A0A0V0GCC5 A0A182IPU3 A0A161MHQ6 A0A131Y097 A0A2M4ATU6 A0A182M5Q7 A0A182VIC8 A0A182NKM1 A0A182TD06 A0A182WJK4 A0A182KQW3 A0A182WZ28 Q7PF81 A0A182RU03 A0A182I2X8
Pubmed
26354079
28756777
23622113
26227816
22118469
18362917
+ More
19820115 17994087 17550304 25315136 15632085 21347285 22936249 10731132 12537572 15975907 20798317 30249741 26108605 26369729 24495485 12537568 12537573 12537574 16110336 17569856 17569867 20075255 17510324 25348373 27129103 29209593 25401762 26823975 22516182 23537049 20920257 23761445 24945155 25474469 28797301 25576852 26334808 20966253 12364791 14747013 17210077
19820115 17994087 17550304 25315136 15632085 21347285 22936249 10731132 12537572 15975907 20798317 30249741 26108605 26369729 24495485 12537568 12537573 12537574 16110336 17569856 17569867 20075255 17510324 25348373 27129103 29209593 25401762 26823975 22516182 23537049 20920257 23761445 24945155 25474469 28797301 25576852 26334808 20966253 12364791 14747013 17210077
EMBL
DQ443233
ABF51322.1
ODYU01009364
SOQ53739.1
KQ460398
KPJ15016.1
+ More
KQ459302 KPJ01811.1 GDQN01002915 JAT88139.1 KZ149900 PZC78588.1 NWSH01000887 PCG73685.1 GAIX01005509 JAA87051.1 JTDY01001572 KOB73490.1 AGBW02010077 OWR49207.1 KQ971372 EFA10351.1 LJIG01022758 KRT78877.1 CH940648 EDW61663.1 CH902619 EDV38122.1 CP012524 ALC42174.1 GBYB01001002 GBYB01001003 GBYB01014882 JAG70769.1 JAG70770.1 JAG84649.1 CM000157 EDW89075.1 CH964282 EDW85559.1 CH933808 EDW08943.1 CH916367 EDW01996.1 CM000071 EAL24719.1 CH479181 EDW31396.1 ADTU01011943 CM002911 KMY91774.1 CH480816 EDW46646.1 CH954177 EDV59799.1 AE013599 AY070538 AAL48009.1 AAM70816.1 CCAG010011030 JXJN01024675 OUUW01000001 SPP74183.1 GL448390 EFN84697.1 QOIP01000008 RLU19841.1 GEHC01000299 JAV47346.1 JRES01000975 KNC26596.1 CM000362 EDX05903.1 GDAI01001728 JAI15875.1 KK115914 KFM66358.1 GAMC01015049 JAB91506.1 KRJ98162.1 KRF80127.1 KMY91775.1 AFH07906.1 GADI01001631 JAA72177.1 ABJB010363074 ABJB010682213 DS795088 EEC10354.1 KRG04378.1 AAZX01009933 CH477272 EAT45255.1 GDHF01005621 GDHF01004176 JAI46693.1 JAI48138.1 GAKP01003175 GAKP01003173 GAKP01003169 JAC55783.1 GEDC01026910 JAS10388.1 GDKW01002033 JAI54562.1 GBBM01002444 JAC32974.1 GFAA01001631 JAU01804.1 KQ434998 KZC13242.1 GBHO01012074 GBRD01003664 GDHC01014031 JAG31530.1 JAG62157.1 JAQ04598.1 APGK01018751 BT127693 KB740082 KB631720 AEE62655.1 ENN81594.1 ERL85578.1 GFDL01012417 JAV22628.1 GFTR01003877 JAW12549.1 KQ981727 KYN36980.1 GFDL01012416 JAV22629.1 DS232080 EDS34190.1 ACPB03018355 ADMH02000578 ETN65806.1 KK854044 PTY10175.1 GGFM01004606 MBW25357.1 GGFK01010686 MBW44007.1 GGFJ01009154 MBW58295.1 GAPW01004066 JAC09532.1 GBBI01002307 JAC16405.1 KQ435922 KOX68434.1 AXCN02000551 GFPF01006651 MAA17797.1 GACK01005861 JAA59173.1 GBGD01002767 JAC86122.1 GECL01000410 JAP05714.1 GEMB01002214 JAS00966.1 GEFM01003976 JAP71820.1 GGFK01010687 MBW44008.1 AXCM01004717 AAAB01008807 EAA45424.1 APCN01000188
KQ459302 KPJ01811.1 GDQN01002915 JAT88139.1 KZ149900 PZC78588.1 NWSH01000887 PCG73685.1 GAIX01005509 JAA87051.1 JTDY01001572 KOB73490.1 AGBW02010077 OWR49207.1 KQ971372 EFA10351.1 LJIG01022758 KRT78877.1 CH940648 EDW61663.1 CH902619 EDV38122.1 CP012524 ALC42174.1 GBYB01001002 GBYB01001003 GBYB01014882 JAG70769.1 JAG70770.1 JAG84649.1 CM000157 EDW89075.1 CH964282 EDW85559.1 CH933808 EDW08943.1 CH916367 EDW01996.1 CM000071 EAL24719.1 CH479181 EDW31396.1 ADTU01011943 CM002911 KMY91774.1 CH480816 EDW46646.1 CH954177 EDV59799.1 AE013599 AY070538 AAL48009.1 AAM70816.1 CCAG010011030 JXJN01024675 OUUW01000001 SPP74183.1 GL448390 EFN84697.1 QOIP01000008 RLU19841.1 GEHC01000299 JAV47346.1 JRES01000975 KNC26596.1 CM000362 EDX05903.1 GDAI01001728 JAI15875.1 KK115914 KFM66358.1 GAMC01015049 JAB91506.1 KRJ98162.1 KRF80127.1 KMY91775.1 AFH07906.1 GADI01001631 JAA72177.1 ABJB010363074 ABJB010682213 DS795088 EEC10354.1 KRG04378.1 AAZX01009933 CH477272 EAT45255.1 GDHF01005621 GDHF01004176 JAI46693.1 JAI48138.1 GAKP01003175 GAKP01003173 GAKP01003169 JAC55783.1 GEDC01026910 JAS10388.1 GDKW01002033 JAI54562.1 GBBM01002444 JAC32974.1 GFAA01001631 JAU01804.1 KQ434998 KZC13242.1 GBHO01012074 GBRD01003664 GDHC01014031 JAG31530.1 JAG62157.1 JAQ04598.1 APGK01018751 BT127693 KB740082 KB631720 AEE62655.1 ENN81594.1 ERL85578.1 GFDL01012417 JAV22628.1 GFTR01003877 JAW12549.1 KQ981727 KYN36980.1 GFDL01012416 JAV22629.1 DS232080 EDS34190.1 ACPB03018355 ADMH02000578 ETN65806.1 KK854044 PTY10175.1 GGFM01004606 MBW25357.1 GGFK01010686 MBW44007.1 GGFJ01009154 MBW58295.1 GAPW01004066 JAC09532.1 GBBI01002307 JAC16405.1 KQ435922 KOX68434.1 AXCN02000551 GFPF01006651 MAA17797.1 GACK01005861 JAA59173.1 GBGD01002767 JAC86122.1 GECL01000410 JAP05714.1 GEMB01002214 JAS00966.1 GEFM01003976 JAP71820.1 GGFK01010687 MBW44008.1 AXCM01004717 AAAB01008807 EAA45424.1 APCN01000188
Proteomes
UP000053240
UP000053268
UP000218220
UP000037510
UP000007151
UP000007266
+ More
UP000192223 UP000091820 UP000008792 UP000007801 UP000092553 UP000002282 UP000095301 UP000007798 UP000009192 UP000001070 UP000001819 UP000008744 UP000005205 UP000192221 UP000001292 UP000008711 UP000000803 UP000095300 UP000092444 UP000092443 UP000078200 UP000092460 UP000268350 UP000008237 UP000279307 UP000037069 UP000000304 UP000054359 UP000001555 UP000002358 UP000005203 UP000008820 UP000076502 UP000019118 UP000030742 UP000075881 UP000078541 UP000002320 UP000015103 UP000000673 UP000069272 UP000053105 UP000075886 UP000075880 UP000075883 UP000075903 UP000075884 UP000075902 UP000075920 UP000075882 UP000076407 UP000007062 UP000075900 UP000075840
UP000192223 UP000091820 UP000008792 UP000007801 UP000092553 UP000002282 UP000095301 UP000007798 UP000009192 UP000001070 UP000001819 UP000008744 UP000005205 UP000192221 UP000001292 UP000008711 UP000000803 UP000095300 UP000092444 UP000092443 UP000078200 UP000092460 UP000268350 UP000008237 UP000279307 UP000037069 UP000000304 UP000054359 UP000001555 UP000002358 UP000005203 UP000008820 UP000076502 UP000019118 UP000030742 UP000075881 UP000078541 UP000002320 UP000015103 UP000000673 UP000069272 UP000053105 UP000075886 UP000075880 UP000075883 UP000075903 UP000075884 UP000075902 UP000075920 UP000075882 UP000076407 UP000007062 UP000075900 UP000075840
Pfam
PF03637 Mob1_phocein
SUPFAM
SSF101152
SSF101152
Gene 3D
ProteinModelPortal
Q1HQ19
A0A2H1WL05
A0A194RGF7
A0A194QEB9
A0A1E1WM92
A0A2W1BU66
+ More
A0A2A4JNP9 S4P5J4 A0A0L7LDL2 A0A212F646 D6X397 A0A0T6AV83 A0A1W4XIX0 A0A1A9W102 B4LK09 B3MIP6 A0A0M5IXV4 A0A0C9QK69 B4P101 A0A1I8MQE3 B4NMH0 B4KNI4 B4J6J5 Q292Z2 B4GC78 A0A158NCJ4 A0A1W4V1Z3 A0A0J9R7W7 B4HQA7 B3NB34 Q7K0E3 A0A1I8P7Q1 A0A1B0FC86 A0A1A9YC02 A0A1A9UU23 A0A1B0AKE4 A0A3B0J442 E2BHY1 A0A3L8DHM0 A0A1W7R880 A0A0L0C2V7 B4QDB6 A0A0K8TPQ0 A0A087TML9 W8BEA4 A0A1W4VEA4 A0A0R1DQY9 A0A0Q9WFL3 A0A0J9TUE8 A0A0B4K6X9 A0A0K8RM50 B7PUT2 A0A0Q9XEW9 K7JC07 A0A088AVR4 Q17F92 A0A0K8WAB8 A0A034WPW0 A0A1B6CA79 A0A0P4VQ69 A0A023GGH9 A0A1E1XRB6 A0A154PN19 A0A0A9YPS0 J3JWS5 A0A182KC29 A0A1Q3F580 A0A224XWX8 A0A195F8P7 A0A1Q3F524 B0WT47 T1IBN4 W5JNT7 A0A2R7VW68 A0A2M3Z9Z1 A0A2M4ATB5 A0A182FG47 A0A2M4BYY2 A0A023EK78 A0A023F4F2 A0A0N0BC34 A0A182QEA4 A0A224YJH1 L7M7P3 A0A069DR44 A0A0V0GCC5 A0A182IPU3 A0A161MHQ6 A0A131Y097 A0A2M4ATU6 A0A182M5Q7 A0A182VIC8 A0A182NKM1 A0A182TD06 A0A182WJK4 A0A182KQW3 A0A182WZ28 Q7PF81 A0A182RU03 A0A182I2X8
A0A2A4JNP9 S4P5J4 A0A0L7LDL2 A0A212F646 D6X397 A0A0T6AV83 A0A1W4XIX0 A0A1A9W102 B4LK09 B3MIP6 A0A0M5IXV4 A0A0C9QK69 B4P101 A0A1I8MQE3 B4NMH0 B4KNI4 B4J6J5 Q292Z2 B4GC78 A0A158NCJ4 A0A1W4V1Z3 A0A0J9R7W7 B4HQA7 B3NB34 Q7K0E3 A0A1I8P7Q1 A0A1B0FC86 A0A1A9YC02 A0A1A9UU23 A0A1B0AKE4 A0A3B0J442 E2BHY1 A0A3L8DHM0 A0A1W7R880 A0A0L0C2V7 B4QDB6 A0A0K8TPQ0 A0A087TML9 W8BEA4 A0A1W4VEA4 A0A0R1DQY9 A0A0Q9WFL3 A0A0J9TUE8 A0A0B4K6X9 A0A0K8RM50 B7PUT2 A0A0Q9XEW9 K7JC07 A0A088AVR4 Q17F92 A0A0K8WAB8 A0A034WPW0 A0A1B6CA79 A0A0P4VQ69 A0A023GGH9 A0A1E1XRB6 A0A154PN19 A0A0A9YPS0 J3JWS5 A0A182KC29 A0A1Q3F580 A0A224XWX8 A0A195F8P7 A0A1Q3F524 B0WT47 T1IBN4 W5JNT7 A0A2R7VW68 A0A2M3Z9Z1 A0A2M4ATB5 A0A182FG47 A0A2M4BYY2 A0A023EK78 A0A023F4F2 A0A0N0BC34 A0A182QEA4 A0A224YJH1 L7M7P3 A0A069DR44 A0A0V0GCC5 A0A182IPU3 A0A161MHQ6 A0A131Y097 A0A2M4ATU6 A0A182M5Q7 A0A182VIC8 A0A182NKM1 A0A182TD06 A0A182WJK4 A0A182KQW3 A0A182WZ28 Q7PF81 A0A182RU03 A0A182I2X8
PDB
5YF4
E-value=1.05104e-72,
Score=692
Ontologies
GO
PANTHER
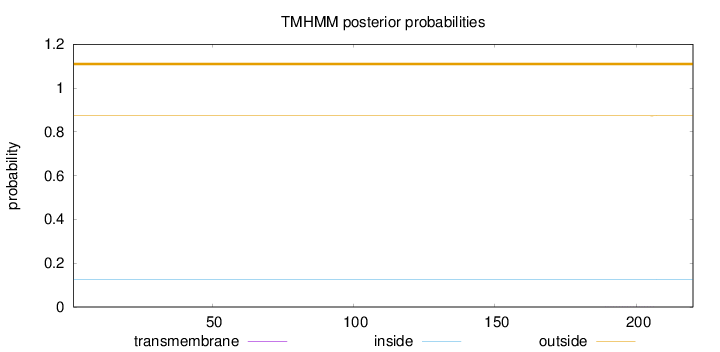
Topology
Length:
220
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00575
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12692
outside
1 - 220
Population Genetic Test Statistics
Pi
128.885625
Theta
120.161038
Tajima's D
0.814433
CLR
6.901641
CSRT
0.606669666516674
Interpretation
Uncertain
