Gene
KWMTBOMO05752
Pre Gene Modal
BGIBMGA006885
Annotation
PREDICTED:_atrial_natriuretic_peptide-converting_enzyme?_partial_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.748 PlasmaMembrane Reliability : 1.237
Sequence
CDS
ATGTTTATTAGTAAACATGATTTCTGTAAAGGCGGTACGAACCGGGGACATTCTACCCGTCACCGAGTAAAGGCCAGACCCGAATGGCCTTGCCACACTCGCCGCAACAAGTGCACTAAATATCACAGACTTGATATGTATTTATTCGGTGACCCTCTGAATAAGCAAACGTTCCGTGGGTCTTTCATCATAAAAAGTAACGCAGAAGACATTCAAAGTTGGAAGGAAAGTAACCAGACTACAGAAAATGCATTTCAGCACATATTATTTCAAATCTATCAGAACTCCGAACTACAATCTTGTTTTGTATCTGCTGACGTATTAGCTTTAGACAGCACAGAGGAAAGGCGGGATGGTATCCGTATACACTTCGAAGTATCCTTCGAGCCTATATTTACTGCGGTAACGACGTCTGAGGTGACAGCCGTACTCGCTCGCGAGTTGACTTCGCCGACAACTTATTTTAGCCGGTACACTACGTATCCAGAAACATTACATATAGAGGAGAATTACATGATTGCATCTTTGCAATTGAAGGAAGACGAAACGACCACATTAGAACCACCTACAACTGAAATTATAAGTTCTGAACCTGAAGTGGAACGACGCGAATGTGTACCGGTTACTTTGGGATTATGTTCACACTTACCTTACAATACCACCTCTTATCCAAATATGGTGGGGCATTCGGACAAAGATGCTCTTATGCGAGACCTCGTGGCGTTTAGAGAGCTTTTAGACGCTGAATGCTCTCACCTTGCTCAGGATTTCGTATGTCAAATGCTACAGCCTCGTTGCGAAGGTAATCATGTAGTGCGACCGTGTCGCTCCTACTGTCGAGCTTTCCACGAAGGCTGCGGTGCTCGCCTACCTGAACGACTAAAAGCACATTTCGACTGTGCGCGATTTCCAGACTTTTTTGGGATCGGATCTTGTGCTCCACAACCAGACTGTCATAGTGATTTACAACGGCTAGCTTTATCGCGGCGAGCGTGTGATAGTATTCCTGACTGCGCCGACGCAGCTGATGAGCGCGAATGTGCGTACTGCAAGAGCGCGGGGGCCGGCGCCCTACGATGTGCCCTTAACGAGCGATGTCTGCCTCCCGCCTTACGTTGCGACGGAAATCCGGACTGCCCCGACGGAAGCGATGAGGCTGGATGTCTGTGGATTTCTCGATCCCTGTCTTCATGGCAACGGGAAAATTCTGAAACAACATTGGGAGCTGTAAGGAATCGCGCCGGATACGCTCTCTGGGCTGAGCGTGGAAGATATGGAAAAATATGTGCTACTCCGTTTGAAAACGATAAAAAAGCTCTTAAAAGCGTTGCCTCATCTTTATGCAAAGCACTTACGTTCACATCATCAATAAGCGCAGAGGCGGTTCTTGATGCAGAGGAATTAAATAAAACAAACTTCGAGAACTCAACAGAATTTAGGTCGATTGCAGAATACGTGGAAATAGTAGATCCCTTTGCTGCTGAAATATCTTTCGTCAAGAGTGAATGTCCGCAGCGAAAAGTCATTAGAATTATTTGTGACCAACTTGATTGTGGAATACCATCAGCAAGAGGCACGAGTTCGACGAGTGGAGTTGACAGCTTACCCCGTTCGGCTCGGCCGGGAGACTGGCCTTGGCAAGCTTCGTTGTTGAGATCTAACATCCATGCTTGCGATGGCTCACTTATACACCCTTCGTGGCTTATAACTACAGCGTCATGTTTTCAGGGGCAACCAAAAGCAGAATGGACAGCCAGATTTGGCACAGTCCGAATACAAAGTACGACACCATGGCAGCAAGAACGTCGTATCGTCGGAATGGTACGATCTCCCGTTGAAGGTAGCATGCTGGTACTGGTTAGACTTGAAGAGCCCGTTGAAATGACCGATTTTGTGCGACCAATATGTCTATCGAATAAGGCCCTTTATAGTGAAGAACGAAGTGTTTGTAACACACTTGGATGGACAAGAAATAGAGACCAGCTGCAAAGAGTTCACATAGTTCCCAGTCCAATGAGCACTTGTGAAAACGTCAGTATAGCCAATCCAAACGGCATCTGCTCGGAACCTTTGTACGACCAAGATGACTGTGACGAAGAAGAATATGCTGGAAGTTCGATGATGTGTTTTGATGATAAAACGAAGCGTTGGTCAATAATGGGTGTCAGTGCATGGCGCATAGCTTGCGCAAAGATAGGTCTAGGGAGGCCTCGCATATACGACCCGGTCACATCACACGTTGATTGGATCCGCAGAACAGTTTCTGATTCAGCAAGATGA
Protein
MFISKHDFCKGGTNRGHSTRHRVKARPEWPCHTRRNKCTKYHRLDMYLFGDPLNKQTFRGSFIIKSNAEDIQSWKESNQTTENAFQHILFQIYQNSELQSCFVSADVLALDSTEERRDGIRIHFEVSFEPIFTAVTTSEVTAVLARELTSPTTYFSRYTTYPETLHIEENYMIASLQLKEDETTTLEPPTTEIISSEPEVERRECVPVTLGLCSHLPYNTTSYPNMVGHSDKDALMRDLVAFRELLDAECSHLAQDFVCQMLQPRCEGNHVVRPCRSYCRAFHEGCGARLPERLKAHFDCARFPDFFGIGSCAPQPDCHSDLQRLALSRRACDSIPDCADAADERECAYCKSAGAGALRCALNERCLPPALRCDGNPDCPDGSDEAGCLWISRSLSSWQRENSETTLGAVRNRAGYALWAERGRYGKICATPFENDKKALKSVASSLCKALTFTSSISAEAVLDAEELNKTNFENSTEFRSIAEYVEIVDPFAAEISFVKSECPQRKVIRIICDQLDCGIPSARGTSSTSGVDSLPRSARPGDWPWQASLLRSNIHACDGSLIHPSWLITTASCFQGQPKAEWTARFGTVRIQSTTPWQQERRIVGMVRSPVEGSMLVLVRLEEPVEMTDFVRPICLSNKALYSEERSVCNTLGWTRNRDQLQRVHIVPSPMSTCENVSIANPNGICSEPLYDQDDCDEEEYAGSSMMCFDDKTKRWSIMGVSAWRIACAKIGLGRPRIYDPVTSHVDWIRRTVSDSAR
Summary
Uniprot
A0A2W1BZC0
A0A2A4J470
A0A2H1WFA3
A0A212EJN8
A0A194RB00
A0A194QEC9
+ More
A0A0L7LPS6 A0A151I8B3 A0A0M9AAB4 A0A0C9RTB9 A0A026WDW7 A0A151J651 A0A088ASV8 A0A154PH37 A0A195BM77 E2AZP0 A0A2J7QDR4 A0A195F7A8 A0A232F4I9 A0A151X415 K7IUE3 A0A0L7R6T8 D6X204 A0A067QW15 F4X7C8 A0A2A3E3F4 A0A1Y1LDQ4 U4U0A1 A0A1V1FKK9 A0A2J7QDQ7 N6UQ81 A0A1B6EE48 A0A1B6FFC0 J9K7S0 A0A1B6LN46 A0A2H8TY26 A0A2S2NDW2 E0V8W5 A0A2S2QD06 A0A0K8TH53 T1H9A8 A0A023F183 A0A158NDF3 A0A1I8MSU2 A0A1I8Q2R9 A0A1B0A310 A0A1B0B9S1 A0A1B0FR42 A0A1A9W403 A0A182S918 A0A1A9XKK7 B4L1M0 A0A182KGY5 A0A0A1WX24 A0A1S4G8N7 W8AKB8 A0A084WPF7 A0A0L0C2I7 A0A1A9UE88 B4M345 A0A0K8W191 A0A182MB77 B4N1I6 A0A182IMI0 A0A182U302 Q7QF48 A0A182FK13 A0A182YH99 A0A182N1E6 A0A182IAM4 A0A0M4EM86 A0A182QQ87 B3MWH5 A0A182VV75 A0A0P8ZKN4 A0A182XBW0 A0A182V2U6 B4JIZ2 A0A182RM70 A0A182P0V2 A0A3B0JXW2 Q9W3H0 Q8MRZ8 A0A1W4VJ06 Q5BIK4 W5JQB3 A0A0R3NZK1 A0A0R1EB26 Q29HR9 B4GYF2 A0A1W4VVR3 A0A0K2VBY2 B4Q0D1 Q16VN8 B3NX20 A0A182LM29 B0WJ26
A0A0L7LPS6 A0A151I8B3 A0A0M9AAB4 A0A0C9RTB9 A0A026WDW7 A0A151J651 A0A088ASV8 A0A154PH37 A0A195BM77 E2AZP0 A0A2J7QDR4 A0A195F7A8 A0A232F4I9 A0A151X415 K7IUE3 A0A0L7R6T8 D6X204 A0A067QW15 F4X7C8 A0A2A3E3F4 A0A1Y1LDQ4 U4U0A1 A0A1V1FKK9 A0A2J7QDQ7 N6UQ81 A0A1B6EE48 A0A1B6FFC0 J9K7S0 A0A1B6LN46 A0A2H8TY26 A0A2S2NDW2 E0V8W5 A0A2S2QD06 A0A0K8TH53 T1H9A8 A0A023F183 A0A158NDF3 A0A1I8MSU2 A0A1I8Q2R9 A0A1B0A310 A0A1B0B9S1 A0A1B0FR42 A0A1A9W403 A0A182S918 A0A1A9XKK7 B4L1M0 A0A182KGY5 A0A0A1WX24 A0A1S4G8N7 W8AKB8 A0A084WPF7 A0A0L0C2I7 A0A1A9UE88 B4M345 A0A0K8W191 A0A182MB77 B4N1I6 A0A182IMI0 A0A182U302 Q7QF48 A0A182FK13 A0A182YH99 A0A182N1E6 A0A182IAM4 A0A0M4EM86 A0A182QQ87 B3MWH5 A0A182VV75 A0A0P8ZKN4 A0A182XBW0 A0A182V2U6 B4JIZ2 A0A182RM70 A0A182P0V2 A0A3B0JXW2 Q9W3H0 Q8MRZ8 A0A1W4VJ06 Q5BIK4 W5JQB3 A0A0R3NZK1 A0A0R1EB26 Q29HR9 B4GYF2 A0A1W4VVR3 A0A0K2VBY2 B4Q0D1 Q16VN8 B3NX20 A0A182LM29 B0WJ26
Pubmed
28756777
22118469
26354079
26227816
24508170
30249741
+ More
20798317 28648823 20075255 18362917 19820115 24845553 21719571 28004739 23537049 28410430 20566863 26823975 25474469 21347285 25315136 17994087 25830018 12364791 24495485 24438588 26108605 25244985 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 15632085 17550304 17510324 20966253
20798317 28648823 20075255 18362917 19820115 24845553 21719571 28004739 23537049 28410430 20566863 26823975 25474469 21347285 25315136 17994087 25830018 12364791 24495485 24438588 26108605 25244985 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 15632085 17550304 17510324 20966253
EMBL
KZ149900
PZC78597.1
NWSH01003259
PCG66669.1
ODYU01008299
SOQ51759.1
+ More
AGBW02014414 OWR41707.1 KQ460398 KPJ15028.1 KQ459302 KPJ01821.1 JTDY01000364 KOB77523.1 KQ978377 KYM94557.1 KQ435700 KOX80437.1 GBYB01010741 JAG80508.1 KK107260 QOIP01000001 EZA54103.1 RLU27640.1 KQ979878 KYN18703.1 KQ434890 KZC10490.1 KQ976438 KYM86861.1 GL444275 EFN61099.1 NEVH01015817 PNF26718.1 KQ981744 KYN36490.1 NNAY01001004 OXU25522.1 KQ982554 KYQ55121.1 AAZX01007282 AAZX01013097 AAZX01016338 KQ414646 KOC66590.1 KQ971371 EFA10739.2 KK852946 KDR13425.1 GL888828 EGI57740.1 KZ288418 PBC26014.1 GEZM01060397 GEZM01060396 JAV71011.1 KB631743 ERL85723.1 FX985319 BAX07332.1 PNF26721.1 APGK01021332 KB740314 ENN80897.1 GEDC01001121 JAS36177.1 GECZ01020868 JAS48901.1 ABLF02032258 GEBQ01014890 JAT25087.1 GFXV01007400 MBW19205.1 GGMR01002741 MBY15360.1 DS234986 EEB09821.1 GGMS01006380 MBY75583.1 GBRD01000992 GDHC01002552 JAG64829.1 JAQ16077.1 ACPB03008383 GBBI01003958 JAC14754.1 ADTU01012389 ADTU01012390 JXJN01010530 CCAG010014821 CCAG010014822 CCAG010014823 CH933810 EDW07656.2 GBXI01011106 JAD03186.1 AAAB01008846 GAMC01020293 GAMC01020292 JAB86262.1 ATLV01025075 KE525369 KFB52101.1 JRES01000984 KNC26442.1 CH940651 EDW65220.1 GDHF01007397 JAI44917.1 AXCM01002730 CH963925 EDW78225.1 EAA06527.5 APCN01001276 CP012528 ALC48784.1 AXCN02002019 CH902625 EDV34960.2 KPU75372.1 KPU75373.1 CH916370 EDV99556.1 OUUW01000011 SPP86894.1 AE014298 AAF46357.2 AFH07291.1 AY119177 AAM51037.1 BT021220 AAX33368.1 ADMH02000772 ETN65095.1 CH379064 KRT06462.1 CM000162 KRK06680.1 EAL31688.2 CH479197 EDW27808.1 HACA01030454 CDW47815.1 EDX02268.1 CH477584 EAT38644.1 CH954180 EDV46849.1 DS231955 EDS28905.1
AGBW02014414 OWR41707.1 KQ460398 KPJ15028.1 KQ459302 KPJ01821.1 JTDY01000364 KOB77523.1 KQ978377 KYM94557.1 KQ435700 KOX80437.1 GBYB01010741 JAG80508.1 KK107260 QOIP01000001 EZA54103.1 RLU27640.1 KQ979878 KYN18703.1 KQ434890 KZC10490.1 KQ976438 KYM86861.1 GL444275 EFN61099.1 NEVH01015817 PNF26718.1 KQ981744 KYN36490.1 NNAY01001004 OXU25522.1 KQ982554 KYQ55121.1 AAZX01007282 AAZX01013097 AAZX01016338 KQ414646 KOC66590.1 KQ971371 EFA10739.2 KK852946 KDR13425.1 GL888828 EGI57740.1 KZ288418 PBC26014.1 GEZM01060397 GEZM01060396 JAV71011.1 KB631743 ERL85723.1 FX985319 BAX07332.1 PNF26721.1 APGK01021332 KB740314 ENN80897.1 GEDC01001121 JAS36177.1 GECZ01020868 JAS48901.1 ABLF02032258 GEBQ01014890 JAT25087.1 GFXV01007400 MBW19205.1 GGMR01002741 MBY15360.1 DS234986 EEB09821.1 GGMS01006380 MBY75583.1 GBRD01000992 GDHC01002552 JAG64829.1 JAQ16077.1 ACPB03008383 GBBI01003958 JAC14754.1 ADTU01012389 ADTU01012390 JXJN01010530 CCAG010014821 CCAG010014822 CCAG010014823 CH933810 EDW07656.2 GBXI01011106 JAD03186.1 AAAB01008846 GAMC01020293 GAMC01020292 JAB86262.1 ATLV01025075 KE525369 KFB52101.1 JRES01000984 KNC26442.1 CH940651 EDW65220.1 GDHF01007397 JAI44917.1 AXCM01002730 CH963925 EDW78225.1 EAA06527.5 APCN01001276 CP012528 ALC48784.1 AXCN02002019 CH902625 EDV34960.2 KPU75372.1 KPU75373.1 CH916370 EDV99556.1 OUUW01000011 SPP86894.1 AE014298 AAF46357.2 AFH07291.1 AY119177 AAM51037.1 BT021220 AAX33368.1 ADMH02000772 ETN65095.1 CH379064 KRT06462.1 CM000162 KRK06680.1 EAL31688.2 CH479197 EDW27808.1 HACA01030454 CDW47815.1 EDX02268.1 CH477584 EAT38644.1 CH954180 EDV46849.1 DS231955 EDS28905.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
UP000078542
+ More
UP000053105 UP000053097 UP000279307 UP000078492 UP000005203 UP000076502 UP000078540 UP000000311 UP000235965 UP000078541 UP000215335 UP000075809 UP000002358 UP000053825 UP000007266 UP000027135 UP000007755 UP000242457 UP000030742 UP000019118 UP000007819 UP000009046 UP000015103 UP000005205 UP000095301 UP000095300 UP000092445 UP000092460 UP000092444 UP000091820 UP000075901 UP000092443 UP000009192 UP000075881 UP000030765 UP000037069 UP000078200 UP000008792 UP000075883 UP000007798 UP000075880 UP000075902 UP000007062 UP000069272 UP000076408 UP000075884 UP000075840 UP000092553 UP000075886 UP000007801 UP000075920 UP000076407 UP000075903 UP000001070 UP000075900 UP000075885 UP000268350 UP000000803 UP000192221 UP000000673 UP000001819 UP000002282 UP000008744 UP000008820 UP000008711 UP000075882 UP000002320
UP000053105 UP000053097 UP000279307 UP000078492 UP000005203 UP000076502 UP000078540 UP000000311 UP000235965 UP000078541 UP000215335 UP000075809 UP000002358 UP000053825 UP000007266 UP000027135 UP000007755 UP000242457 UP000030742 UP000019118 UP000007819 UP000009046 UP000015103 UP000005205 UP000095301 UP000095300 UP000092445 UP000092460 UP000092444 UP000091820 UP000075901 UP000092443 UP000009192 UP000075881 UP000030765 UP000037069 UP000078200 UP000008792 UP000075883 UP000007798 UP000075880 UP000075902 UP000007062 UP000069272 UP000076408 UP000075884 UP000075840 UP000092553 UP000075886 UP000007801 UP000075920 UP000076407 UP000075903 UP000001070 UP000075900 UP000075885 UP000268350 UP000000803 UP000192221 UP000000673 UP000001819 UP000002282 UP000008744 UP000008820 UP000008711 UP000075882 UP000002320
Pfam
Interpro
IPR002172
LDrepeatLR_classA_rpt
+ More
IPR036055 LDL_receptor-like_sf
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR036364 SEA_dom_sf
IPR020067 Frizzled_dom
IPR000082 SEA_dom
IPR036790 Frizzled_dom_sf
IPR023415 LDLR_class-A_CS
IPR004018 RPEL_repeat
IPR017343 UCP037987
IPR011059 Metal-dep_hydrolase_composite
IPR006132 Asp/Orn_carbamoyltranf_P-bd
IPR036901 Asp/Orn_carbamoylTrfase_sf
IPR005480 CarbamoylP_synth_lsu_oligo
IPR017926 GATASE
IPR036480 CarbP_synth_ssu_N_sf
IPR002474 CarbamoylP_synth_ssu_N
IPR036897 CarbamoylP_synth_lsu_oligo_sf
IPR002082 Asp_carbamoyltransf
IPR029062 Class_I_gatase-like
IPR011761 ATP-grasp
IPR006131 Asp_carbamoyltransf_Asp/Orn-bd
IPR016185 PreATP-grasp_dom_sf
IPR006274 CarbamoylP_synth_ssu
IPR035686 CPSase_GATase1
IPR005479 CbamoylP_synth_lsu-like_ATP-bd
IPR006130 Asp/Orn_carbamoylTrfase
IPR036055 LDL_receptor-like_sf
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR036364 SEA_dom_sf
IPR020067 Frizzled_dom
IPR000082 SEA_dom
IPR036790 Frizzled_dom_sf
IPR023415 LDLR_class-A_CS
IPR004018 RPEL_repeat
IPR017343 UCP037987
IPR011059 Metal-dep_hydrolase_composite
IPR006132 Asp/Orn_carbamoyltranf_P-bd
IPR036901 Asp/Orn_carbamoylTrfase_sf
IPR005480 CarbamoylP_synth_lsu_oligo
IPR017926 GATASE
IPR036480 CarbP_synth_ssu_N_sf
IPR002474 CarbamoylP_synth_ssu_N
IPR036897 CarbamoylP_synth_lsu_oligo_sf
IPR002082 Asp_carbamoyltransf
IPR029062 Class_I_gatase-like
IPR011761 ATP-grasp
IPR006131 Asp_carbamoyltransf_Asp/Orn-bd
IPR016185 PreATP-grasp_dom_sf
IPR006274 CarbamoylP_synth_ssu
IPR035686 CPSase_GATase1
IPR005479 CbamoylP_synth_lsu-like_ATP-bd
IPR006130 Asp/Orn_carbamoylTrfase
SUPFAM
ProteinModelPortal
A0A2W1BZC0
A0A2A4J470
A0A2H1WFA3
A0A212EJN8
A0A194RB00
A0A194QEC9
+ More
A0A0L7LPS6 A0A151I8B3 A0A0M9AAB4 A0A0C9RTB9 A0A026WDW7 A0A151J651 A0A088ASV8 A0A154PH37 A0A195BM77 E2AZP0 A0A2J7QDR4 A0A195F7A8 A0A232F4I9 A0A151X415 K7IUE3 A0A0L7R6T8 D6X204 A0A067QW15 F4X7C8 A0A2A3E3F4 A0A1Y1LDQ4 U4U0A1 A0A1V1FKK9 A0A2J7QDQ7 N6UQ81 A0A1B6EE48 A0A1B6FFC0 J9K7S0 A0A1B6LN46 A0A2H8TY26 A0A2S2NDW2 E0V8W5 A0A2S2QD06 A0A0K8TH53 T1H9A8 A0A023F183 A0A158NDF3 A0A1I8MSU2 A0A1I8Q2R9 A0A1B0A310 A0A1B0B9S1 A0A1B0FR42 A0A1A9W403 A0A182S918 A0A1A9XKK7 B4L1M0 A0A182KGY5 A0A0A1WX24 A0A1S4G8N7 W8AKB8 A0A084WPF7 A0A0L0C2I7 A0A1A9UE88 B4M345 A0A0K8W191 A0A182MB77 B4N1I6 A0A182IMI0 A0A182U302 Q7QF48 A0A182FK13 A0A182YH99 A0A182N1E6 A0A182IAM4 A0A0M4EM86 A0A182QQ87 B3MWH5 A0A182VV75 A0A0P8ZKN4 A0A182XBW0 A0A182V2U6 B4JIZ2 A0A182RM70 A0A182P0V2 A0A3B0JXW2 Q9W3H0 Q8MRZ8 A0A1W4VJ06 Q5BIK4 W5JQB3 A0A0R3NZK1 A0A0R1EB26 Q29HR9 B4GYF2 A0A1W4VVR3 A0A0K2VBY2 B4Q0D1 Q16VN8 B3NX20 A0A182LM29 B0WJ26
A0A0L7LPS6 A0A151I8B3 A0A0M9AAB4 A0A0C9RTB9 A0A026WDW7 A0A151J651 A0A088ASV8 A0A154PH37 A0A195BM77 E2AZP0 A0A2J7QDR4 A0A195F7A8 A0A232F4I9 A0A151X415 K7IUE3 A0A0L7R6T8 D6X204 A0A067QW15 F4X7C8 A0A2A3E3F4 A0A1Y1LDQ4 U4U0A1 A0A1V1FKK9 A0A2J7QDQ7 N6UQ81 A0A1B6EE48 A0A1B6FFC0 J9K7S0 A0A1B6LN46 A0A2H8TY26 A0A2S2NDW2 E0V8W5 A0A2S2QD06 A0A0K8TH53 T1H9A8 A0A023F183 A0A158NDF3 A0A1I8MSU2 A0A1I8Q2R9 A0A1B0A310 A0A1B0B9S1 A0A1B0FR42 A0A1A9W403 A0A182S918 A0A1A9XKK7 B4L1M0 A0A182KGY5 A0A0A1WX24 A0A1S4G8N7 W8AKB8 A0A084WPF7 A0A0L0C2I7 A0A1A9UE88 B4M345 A0A0K8W191 A0A182MB77 B4N1I6 A0A182IMI0 A0A182U302 Q7QF48 A0A182FK13 A0A182YH99 A0A182N1E6 A0A182IAM4 A0A0M4EM86 A0A182QQ87 B3MWH5 A0A182VV75 A0A0P8ZKN4 A0A182XBW0 A0A182V2U6 B4JIZ2 A0A182RM70 A0A182P0V2 A0A3B0JXW2 Q9W3H0 Q8MRZ8 A0A1W4VJ06 Q5BIK4 W5JQB3 A0A0R3NZK1 A0A0R1EB26 Q29HR9 B4GYF2 A0A1W4VVR3 A0A0K2VBY2 B4Q0D1 Q16VN8 B3NX20 A0A182LM29 B0WJ26
PDB
5GVT
E-value=2.90631e-19,
Score=237
Ontologies
GO
Topology
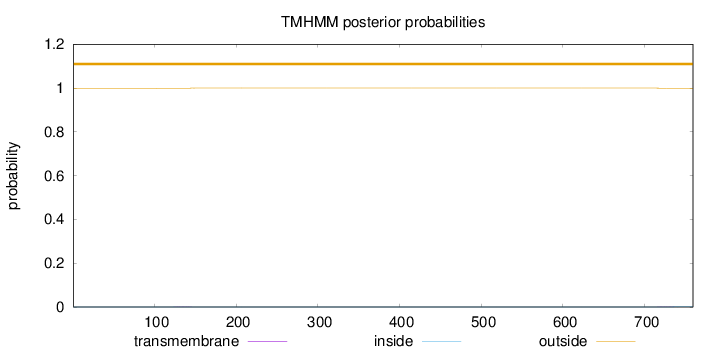
Length:
759
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01992
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00044
outside
1 - 759
Population Genetic Test Statistics
Pi
191.653946
Theta
182.569067
Tajima's D
0.203417
CLR
1.934973
CSRT
0.425578721063947
Interpretation
Uncertain
