Gene
KWMTBOMO05746
Pre Gene Modal
BGIBMGA006882
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_C_member_Sur_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.251
Sequence
CDS
ATGGAGGCAGTTTGTGGTCAAGGTGATAGAAGCTGTGAGATGCGAGCATTGAATGCCGCACTACATTCACTGCACCTGTACTGCGCTACAATCACTGCGGGATTAATGTACTGTTGTTCCACCACGCACAAACATTACAGATATAAGAAGAACTGGAGAGCTCAAAATGTACGAGGAGTGGCCAGTCTTGCTCTGTTTGGTATCTGGTGCTGTGGGGCAGCGGCGGCACTTCTACGTCGCACGGATACTGTACGATTAGTCTCGGCCCTCATGGCACCTCTTGCCTGGATCGCAGCGGCCTGCTTCCACGTCGCTCTATGGAATAGACAATCCTCAGCGCCTATATTGTACTTGGCGATATATTGGTCACTCTCAGCCATATCAGCCGCTTCCATTCTTTGGCAGCACATCCGTGTCGGCGCTGAAGTCAACCATATAGAATTATATATTCAGGGTGCCTCATTGCTGCTTTCTTTAACGATATCTGCCGTGGACTGCGCGTGCTTTTACGAAGTGGTAACAAAACATTTTAGTCATCGCGACAAGACGACGAACAGCTCAAGCAAAGTCTCTTACAAACACAATAGCTCTCATTTTTATTCCAAAATCTCGTTTTATTGGATAAACGGTCTGTTATACAAAGGGTATGAATCTCCATTAGAGATGGAGGATTTAGGAGAATTACCAGATAAAGAAAAATCAGTCAAATATTACAAAAAAATCCACAGCATTTATAAAAGCGTTGAGAAAACAAAGTATAACGATGGCCCTAGTATATGGACATGTTACCTATGTAGAGTGTGGCCAAATTTTTACATTGGTGGTATTTTAAAACTTTTAGGTGACTTAATAGGTTTAGTTCCACCATTGGGGCTGGCTTTCATAATTCAGTTTATTGAAAGTCCGATTAGTAATGAACATAACAGTAACGAAGCACATGTAACTTTAAGAGAATTTATCGGTAATGGCTACGTCATGTTATTAGTAATTACTTTGGCTTTGATTTATCAGGCCTTACTCTCTCAAAATTCGACGCATTTGGTAACAGTGGAAGGTGCCAGATTAAAGACTGCTTTACAAGCTATGATTTATGACAAATGTATGCGTATAGCATCTTGGACGCCATCGGAGTCAAATTTAGACGAAGATTCCCCTTTGCTGATGAATTCGGACGATGAGAGCAATACGCGATCTGGTCTCATAACGAATTTAGTTTCACAAGACACGTACAATATTATGTCTTGCGTTTGGATTTGTCATTACATATGGGCTATACCGCTTAAGGTGGCTGTGATACTATATCTATTGTACATGAAATTAGGCGTAAGCGCAATTATCGGAGCAGTAGCAAGCGTGTTAGTGATCACACCCTTGCAATTTCTTGTAGGAAAGAAACTTTCAGATAACTCTAAAGATATTTCTAAATGTACGGATCACAGAATTTCCAAAATGACCGAAATATTGCAAGGAATAAATGTAATTAAACTCTACGTATGGGAAGATTTGTTTAAAGAAAAAATATTACAACTTAGAGAAGTCGAACTGAAACTGCTAAATAAAGATTCATTATACTGGAGTTTTTTGACTTTTTCGACTCAAGTCTCTACAATCTTAATCACAGTTATTACATTCACTGTATATTTTTTCATAGAAGAAGGAACCGGCTTAACCGCGATGAACGTATTCGCTGGCTTAGCCTTCTTTAATCAGCTTTCCGTTCCATTGCTTATTTTGCCAGTCACTGTCCTCATGATAATTCAAGCTATGGTGAGCACAAGACGAATCAAAGATTTTTTGGATCTTCCCGAATCGAACAATGTCGGAAACGAAATTACTGAATATGAAGAGTTATTGAACAAACCTAATGTCCCTTGTAATGACAATACACATCAGAATGACGTGCCATCTCACGATGTAATGGCCAATAAAGATGAGATGTGTGAAATTGAGTTCGCTGAGCAATTATCAAATGAGTCACACGTTAATAAATCGGAATCCTCTCAATTGGTCACATTTCGAAATGCGGCGTTCACGTGGGGTGTGAAAGATGATCTATTGGAAGTTGGCGATCTTGAAATTCCTACAGGGAAACTAATTATGGTTGTCGGAGGCACAAGTGCTGGAAAATCATCATTCTTGTCTGCTTTAATTGGAGAAATGTATCTTGAAAGAGGAGATGTGTTATATAATCGAAACAGTTCGAAGTGGTATGCCGGTCAGCCTCCGTGGTTGCTAGAGGCTTCAGTTCGCGATAACATAATAATGGGTCATTCTTGGTGTCAGAAAAATTATGCACGTGTTCTACGCGCCACAGGTCTGCGGCCAGACTTACAACTTTTACCAGAAGGCGACGCAACCCGCCTGGGCAGCCACGGTACTCCCTTATCCGGCGGTCAACGCATTCGCGTGAGCATTGCTCGAGCATTATACTCCAAGGCACAACTGCTGGCTTTAGACGAGCCGCTAGCTGCGCTCGACGCTGCACTGGCACGTCATGTGGTAGCGCGAGGACTGGTGCCGGCTGTCCGCGCAGGGCGAACCATTGTCCTCGCAACTAACAGACTGGAACTACTACACTATGCTGACTTGATTATCGCAATGGATAATGGACGTGTGAGCGGATACGGTCGAGTGTCCAGCGCCAGTAACGACAGCTACCTATCGCGCTGGGTGCAGCTTGCAAGCGAAGCTCGTGCAGCTGCAGCGCGAGGATGTGCTGGTCCTCCTGACGGAACTGCACGGGAACGGGCGCGGCTCGTGCGAGTGCTCAGCCGGGCTAAATTCCAAAGATGCGCATCAGAGGATACTGCAATCGGCATAACAGAGGCAGCAGGGGCGCATTTGTTAGCAGAGGTACCGATGTACACTGGCGGTAGTTGGCGTAGAGTAGTGCAGCAACAGAGACCGCCCCTCATGAGGCAGTTCTCATCACCGCCACCATCATCTATATGGCGTCGTGAAATACGTCGTACAGTCAGTGCGGATGAATCAAACACTACACTACAACGAGAAGTCAGTCTTCTTCGACGCTGGCTACGACCACAAGGAAACCGAACTCTTAGGAGATGGACCCCGAGAAATCTAAGACGTTTAATCAGTTCAGAATCCGATACACCTGCCCCGGACACTAATGGAATTAATGAAGAATCCTTAGTGGAGACCACTTACAGCACAGTTAGCATAAACGGAGGGGATACAAATGTCGGAATGATTAATGACGAATCTTCACTAGATAAGAGTCAATCTTGGAATGACGTCAGTGAGTGCAAGATATGGCTAGAGTATGGTCTCGCGTGCGGTTGGTGGGGTATCGCCTTCTGGGTAGCAGCAGCCACAGCGCAAGGCTTAGCGCTGGCAGCCGACTACTGGTTGATGTGCCTCACTAATGACAATGCCACTGAAAACTTTTCTGATGATCAGTTGTGGAGCAGTGTCCGCAAGTACGGGTTGTGGTGCGCTGCGGGGGCGGGGGCGGCGGGCGCGGCTCAGGCGGCGTGCGCATGCGCGGGGGCAGGAGCGCGGCGAGTGCTGCATGAGCGCCTGCTGCACGCCGCGCTCTACGCCAGCCCCCACCACCACCACACTACGCCCACCGGGACCACGCTCCATCGCTTCTCCGCTGACGTTCTATTGCTAGACAAAAAATTACCGACAGCCATAAGTCGGTGGACTCAGTTAGCTCTGCTATGTGCTGCGGCTATGCTTGTCAATGCTGTAGTCGCGCCCTGGAGCCTGACAGCACTCATACCGGCATTCTTGTGTTACCTCTGCCTACAATCTGTTTATTTACGTAATTCTAGGGAGTTGCAGCAAGTGGAAGGACAAAGCGCCGGAAGAGTGGTGTCTCTTTGCAGAGAGACGTGTGCTGGAGGTTCTACTATACGAGCCAGCCGACTACAGACCCGCATGCGGACAGACTTCCTTCGACGTCTCGACTATAATCACAACGCTCTGCTGCTCCTCAATACTGCTAACCGGTGGCTTGGATTGTCTCTGGATCTTGTTGGCGCTGCGAGTGTTTGCGTGTCCCTGTGCGTGTGCATGTACAGCGGCAGTAGAGGCGCGGTCGCGGGACTAGCGGGCGCGTACTCGCTGCTCCTGCCCGCCTATCTCGCTCATCTTGCCAAATGCCGCGCCGACCTCGACCAGCAACTCGCCGCGCTGCATCGTATACATCGTGACACCAATGTACCCCGCGAAGATTATAGAGAGCATTGCTCTATTCCTGCAAATTGGCAAAGAAGCGGAAAAATTGAATTTCAAAATGTCAGCGTGCAGCACGAACCTAACTCGCAGAATATTTTGAACAAAATCAATATATCTATAGCTCCCGGTGAGAAGGTAGCAATATGCGGCAGGAGTGGTAGCGGCAAGTCTACGTTATTAATGACGTGTGTTGGAGCAACCAGTATTGCCGAAGGACGAGTGCTTATCGACGGGCAGGACGTGTCGCGCGTGCCGCTGCGGGCGTTACGTCACCGCGTGGTCGTCCTGCCGCAAGAGCCTGCGATGTTCTCGGGCACGCTGCGAGAAAACCTCGACCCGCTCGCCGTGCACACTGACGAAGAAATATGGCACAGTCTCAAAGTTGTCGGATTATATGATTTCGTCAGTGCTCAATCGGCTGGTTTGGAGTGCAGTATAGCGGGGCGCGGCGGGTGGAGCGCGGGACGGGCGGCGCGTGCGTGTGCGGCCCGCGCGGCGCTGCACGCCCGTTTAGCGGCCGCCCTGTTGCTGGACGAGCCCGCCGCCGCACTCGACGCCGCAGCCGAGCGAGCACTTCTCGCCGCAATGGCGCGCATCGCACCCGACACCACCATCATCACCGTCGCACATCGCATATCGTCAGTGCGCGGGTACGACCGCGCCGTCGTACTGGAGGAGGGTCGCGTGGTGGAGCAGGGCGAGGTGGGCCCGCTGCTGTCGCGGCCGGTCTCACGCCTCGCCCGCATGCTCGCCGCCGCGCACACGCACCTGATCGCCGTGTGTGGAGCGAAGGCGGACCGGAACTGA
Protein
MEAVCGQGDRSCEMRALNAALHSLHLYCATITAGLMYCCSTTHKHYRYKKNWRAQNVRGVASLALFGIWCCGAAAALLRRTDTVRLVSALMAPLAWIAAACFHVALWNRQSSAPILYLAIYWSLSAISAASILWQHIRVGAEVNHIELYIQGASLLLSLTISAVDCACFYEVVTKHFSHRDKTTNSSSKVSYKHNSSHFYSKISFYWINGLLYKGYESPLEMEDLGELPDKEKSVKYYKKIHSIYKSVEKTKYNDGPSIWTCYLCRVWPNFYIGGILKLLGDLIGLVPPLGLAFIIQFIESPISNEHNSNEAHVTLREFIGNGYVMLLVITLALIYQALLSQNSTHLVTVEGARLKTALQAMIYDKCMRIASWTPSESNLDEDSPLLMNSDDESNTRSGLITNLVSQDTYNIMSCVWICHYIWAIPLKVAVILYLLYMKLGVSAIIGAVASVLVITPLQFLVGKKLSDNSKDISKCTDHRISKMTEILQGINVIKLYVWEDLFKEKILQLREVELKLLNKDSLYWSFLTFSTQVSTILITVITFTVYFFIEEGTGLTAMNVFAGLAFFNQLSVPLLILPVTVLMIIQAMVSTRRIKDFLDLPESNNVGNEITEYEELLNKPNVPCNDNTHQNDVPSHDVMANKDEMCEIEFAEQLSNESHVNKSESSQLVTFRNAAFTWGVKDDLLEVGDLEIPTGKLIMVVGGTSAGKSSFLSALIGEMYLERGDVLYNRNSSKWYAGQPPWLLEASVRDNIIMGHSWCQKNYARVLRATGLRPDLQLLPEGDATRLGSHGTPLSGGQRIRVSIARALYSKAQLLALDEPLAALDAALARHVVARGLVPAVRAGRTIVLATNRLELLHYADLIIAMDNGRVSGYGRVSSASNDSYLSRWVQLASEARAAAARGCAGPPDGTARERARLVRVLSRAKFQRCASEDTAIGITEAAGAHLLAEVPMYTGGSWRRVVQQQRPPLMRQFSSPPPSSIWRREIRRTVSADESNTTLQREVSLLRRWLRPQGNRTLRRWTPRNLRRLISSESDTPAPDTNGINEESLVETTYSTVSINGGDTNVGMINDESSLDKSQSWNDVSECKIWLEYGLACGWWGIAFWVAAATAQGLALAADYWLMCLTNDNATENFSDDQLWSSVRKYGLWCAAGAGAAGAAQAACACAGAGARRVLHERLLHAALYASPHHHHTTPTGTTLHRFSADVLLLDKKLPTAISRWTQLALLCAAAMLVNAVVAPWSLTALIPAFLCYLCLQSVYLRNSRELQQVEGQSAGRVVSLCRETCAGGSTIRASRLQTRMRTDFLRRLDYNHNALLLLNTANRWLGLSLDLVGAASVCVSLCVCMYSGSRGAVAGLAGAYSLLLPAYLAHLAKCRADLDQQLAALHRIHRDTNVPREDYREHCSIPANWQRSGKIEFQNVSVQHEPNSQNILNKINISIAPGEKVAICGRSGSGKSTLLMTCVGATSIAEGRVLIDGQDVSRVPLRALRHRVVVLPQEPAMFSGTLRENLDPLAVHTDEEIWHSLKVVGLYDFVSAQSAGLECSIAGRGGWSAGRAARACAARAALHARLAAALLLDEPAAALDAAAERALLAAMARIAPDTTIITVAHRISSVRGYDRAVVLEEGRVVEQGEVGPLLSRPVSRLARMLAAAHTHLIAVCGAKADRN
Summary
Uniprot
Pubmed
EMBL
ODYU01009012
SOQ53094.1
NWSH01003563
PCG66110.1
KQ459302
KPJ01828.1
+ More
GBYB01004583 JAG74350.1 GL452292 EFN77618.1 QOIP01000008 RLU19415.1 KK107088 EZA59771.1 GL438684 EFN68626.1 GL764129 EFZ18436.1 KZ288198 PBC33676.1 GDHC01005285 JAQ13344.1 PYGN01001043 PSN38190.1 KQ435922 KOX68441.1 GECZ01011150 JAS58619.1 KQ982934 KYQ49178.1 KQ978076 KYM97235.1 KQ978619 KYN29755.1 KQ976396 KYM92924.1 ADTU01011879 ADTU01011880 ADTU01011881 GECZ01023725 JAS46044.1 GFXV01005038 MBW16843.1 GGMS01014904 MBY84107.1 GECZ01007393 JAS62376.1 GL888417 EGI61249.1 ABLF02035352 GGMR01017791 MBY30410.1 GFAC01002146 JAT97042.1
GBYB01004583 JAG74350.1 GL452292 EFN77618.1 QOIP01000008 RLU19415.1 KK107088 EZA59771.1 GL438684 EFN68626.1 GL764129 EFZ18436.1 KZ288198 PBC33676.1 GDHC01005285 JAQ13344.1 PYGN01001043 PSN38190.1 KQ435922 KOX68441.1 GECZ01011150 JAS58619.1 KQ982934 KYQ49178.1 KQ978076 KYM97235.1 KQ978619 KYN29755.1 KQ976396 KYM92924.1 ADTU01011879 ADTU01011880 ADTU01011881 GECZ01023725 JAS46044.1 GFXV01005038 MBW16843.1 GGMS01014904 MBY84107.1 GECZ01007393 JAS62376.1 GL888417 EGI61249.1 ABLF02035352 GGMR01017791 MBY30410.1 GFAC01002146 JAT97042.1
Proteomes
Interpro
IPR003439
ABC_transporter-like
+ More
IPR036640 ABC1_TM_sf
IPR011527 ABC1_TM_dom
IPR027417 P-loop_NTPase
IPR003593 AAA+_ATPase
IPR018488 cNMP-bd_CS
IPR000595 cNMP-bd_dom
IPR035014 STKc_cGK
IPR000719 Prot_kinase_dom
IPR000961 AGC-kinase_C
IPR018490 cNMP-bd-like
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR014710 RmlC-like_jellyroll
IPR017871 ABC_transporter_CS
IPR002374 cGMP_dep_kinase
IPR017441 Protein_kinase_ATP_BS
IPR036640 ABC1_TM_sf
IPR011527 ABC1_TM_dom
IPR027417 P-loop_NTPase
IPR003593 AAA+_ATPase
IPR018488 cNMP-bd_CS
IPR000595 cNMP-bd_dom
IPR035014 STKc_cGK
IPR000719 Prot_kinase_dom
IPR000961 AGC-kinase_C
IPR018490 cNMP-bd-like
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR014710 RmlC-like_jellyroll
IPR017871 ABC_transporter_CS
IPR002374 cGMP_dep_kinase
IPR017441 Protein_kinase_ATP_BS
Gene 3D
ProteinModelPortal
PDB
6C3P
E-value=2.88839e-65,
Score=637
Ontologies
GO
Topology
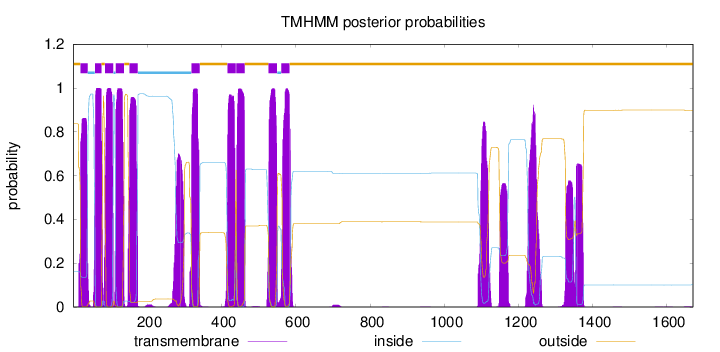
Length:
1670
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
313.94773
Exp number, first 60 AAs:
20.92026
Total prob of N-in:
0.16276
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 39
inside
40 - 58
TMhelix
59 - 76
outside
77 - 85
TMhelix
86 - 108
inside
109 - 114
TMhelix
115 - 137
outside
138 - 151
TMhelix
152 - 174
inside
175 - 318
TMhelix
319 - 341
outside
342 - 415
TMhelix
416 - 438
inside
439 - 439
TMhelix
440 - 462
outside
463 - 526
TMhelix
527 - 549
inside
550 - 560
TMhelix
561 - 583
outside
584 - 1670
Population Genetic Test Statistics
Pi
220.57997
Theta
181.411935
Tajima's D
0.449946
CLR
0.221259
CSRT
0.500574971251437
Interpretation
Uncertain
