Gene
KWMTBOMO05742 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006878
Annotation
PREDICTED:_integrin_beta_subunit_1_isoform_X1_[Bombyx_mori]
Full name
Integrin beta
Location in the cell
Extracellular Reliability : 3.775
Sequence
CDS
ATGTACATAAAACGAACGAGTTTATTGCTGGCAAGCGTGTGGGCGAGCCTGTTGGTCGTGTGTTTTACGCAACACGCAGAGCAGCTGCTTGCCCAGAATCCTTGCTCGAGTAAAACGACCTGTAGCGAATGCGCTCGGACTCCCAGCTGTGCGTGGTGCTTCGCCACCGACTTCAATGGACCTCGATGTTTTAATCCCGCAATGGAAGGCGGTACCGGTGGTTGCGATGAAGCCTACATTTTCAATCCCGACAATCAGCTGTTTGTCGACCCTGCATATAACAGAGACTTGAGTAGAGCAAAGGGTCGCATGGGAATGGCAACAGGCTCGGCAACTTACGAAGAGTCAATGAGCTCTAAGGGAAGTTCATTTGCTGGCAGCGGGAGTGCCGCTGGTGCAGCTGGTGCTGGCGGGAATCTCGTACAAATGAAGCCCCAGAGAGTGAAGCTACAGATGAGAATGAGTATGTCTTCTTCTTTTTCTTGTCCTTGTCCTATGTGTGGTCAGTGCAGCATGTTCACTTTTACCATTCAACTCTACTTAGTCTGGCCATAA
Protein
MYIKRTSLLLASVWASLLVVCFTQHAEQLLAQNPCSSKTTCSECARTPSCAWCFATDFNGPRCFNPAMEGGTGGCDEAYIFNPDNQLFVDPAYNRDLSRAKGRMGMATGSATYEESMSSKGSSFAGSGSAAGAAGAGGNLVQMKPQRVKLQMRMSMSSSFSCPCPMCGQCSMFTFTIQLYLVWP
Summary
Similarity
Belongs to the integrin beta chain family.
Feature
chain Integrin beta
Uniprot
H9JBI3
A0A076JVB8
D1MBK3
A0A3S2P1P8
S4PK99
Q86G85
+ More
A2TIK8 C6KE04 D2DMM8 A0A2A4K6E0 A0A023UG44 A0A1E1WNR2 A0A2H1WGL4 A0A194PEH3 A0A194RGI2 F8TAD6 A0A1W4XHH0 A0A1W4XS08 D6X0D6 V5G6J5 V5GVJ5 A0A1B6ET25 A0A1B6FVP7 A0A1B6JXH3 A0A1B0AX62 A0A154PMC2 A0A2P8Y2E2 E2A6Z5 A0A2P9A990 A0A336MWD6 B4NEJ2 A0A1J1HRB8 A0A1B6MUI8 A0A0P5CDN4 A0A0N8A8C6 E9GI17 A0A336MBS4 A0A336M8L1 A0A1I8PFU3 A0A0P6IR12 A0A164YYE1 A0A0P5IU01 A0A0N8B5P6 A0A0P4Y6Q1 A0A0P6AIE1
A2TIK8 C6KE04 D2DMM8 A0A2A4K6E0 A0A023UG44 A0A1E1WNR2 A0A2H1WGL4 A0A194PEH3 A0A194RGI2 F8TAD6 A0A1W4XHH0 A0A1W4XS08 D6X0D6 V5G6J5 V5GVJ5 A0A1B6ET25 A0A1B6FVP7 A0A1B6JXH3 A0A1B0AX62 A0A154PMC2 A0A2P8Y2E2 E2A6Z5 A0A2P9A990 A0A336MWD6 B4NEJ2 A0A1J1HRB8 A0A1B6MUI8 A0A0P5CDN4 A0A0N8A8C6 E9GI17 A0A336MBS4 A0A336M8L1 A0A1I8PFU3 A0A0P6IR12 A0A164YYE1 A0A0P5IU01 A0A0N8B5P6 A0A0P4Y6Q1 A0A0P6AIE1
Pubmed
EMBL
BABH01030933
BABH01030934
KJ511854
AII79413.1
GU129932
ACY74431.1
+ More
RSAL01000053 RVE50105.1 GAIX01004620 JAA87940.1 AY237588 AAO85806.1 EF202843 ABM91320.1 GQ178290 ACS66819.1 FJ599745 ACU32665.1 NWSH01000130 PCG79230.1 KJ462516 AHY00655.1 GDQN01007634 GDQN01002429 JAT83420.1 JAT88625.1 ODYU01008525 SOQ52209.1 KQ459606 KPI91428.1 KQ460398 KPJ15041.1 HQ829425 ADW20295.1 KQ971372 EFA10689.2 GALX01002761 JAB65705.1 GALX01002759 JAB65707.1 GECZ01028645 JAS41124.1 GECZ01015487 JAS54282.1 GECU01003808 JAT03899.1 JXJN01005087 JXJN01005088 KQ434978 KZC13006.1 PYGN01001024 PSN38364.1 GL437208 EFN70798.1 LT717638 SIW61630.1 UFQT01003187 SSX34712.1 CH964239 EDW82161.2 CVRI01000020 CRK90591.1 GEBQ01000406 JAT39571.1 GDIP01172638 JAJ50764.1 GDIP01172529 JAJ50873.1 GL732545 EFX80954.1 UFQT01000673 SSX26379.1 SSX26380.1 GDIQ01007080 JAN87657.1 LRGB01000868 KZS15734.1 GDIP01253007 GDIQ01269714 GDIQ01227795 GDIQ01227793 GDIQ01225644 GDIQ01210290 GDIQ01191691 JAI70394.1 JAK41435.1 GDIQ01245981 GDIQ01227794 GDIQ01210291 GDIQ01207807 GDIQ01206458 GDIQ01203248 GDIQ01191690 GDIQ01132238 GDIQ01113476 GDIP01089236 JAK41434.1 JAM14479.1 GDIP01231880 JAI91521.1 GDIP01030205 JAM73510.1
RSAL01000053 RVE50105.1 GAIX01004620 JAA87940.1 AY237588 AAO85806.1 EF202843 ABM91320.1 GQ178290 ACS66819.1 FJ599745 ACU32665.1 NWSH01000130 PCG79230.1 KJ462516 AHY00655.1 GDQN01007634 GDQN01002429 JAT83420.1 JAT88625.1 ODYU01008525 SOQ52209.1 KQ459606 KPI91428.1 KQ460398 KPJ15041.1 HQ829425 ADW20295.1 KQ971372 EFA10689.2 GALX01002761 JAB65705.1 GALX01002759 JAB65707.1 GECZ01028645 JAS41124.1 GECZ01015487 JAS54282.1 GECU01003808 JAT03899.1 JXJN01005087 JXJN01005088 KQ434978 KZC13006.1 PYGN01001024 PSN38364.1 GL437208 EFN70798.1 LT717638 SIW61630.1 UFQT01003187 SSX34712.1 CH964239 EDW82161.2 CVRI01000020 CRK90591.1 GEBQ01000406 JAT39571.1 GDIP01172638 JAJ50764.1 GDIP01172529 JAJ50873.1 GL732545 EFX80954.1 UFQT01000673 SSX26379.1 SSX26380.1 GDIQ01007080 JAN87657.1 LRGB01000868 KZS15734.1 GDIP01253007 GDIQ01269714 GDIQ01227795 GDIQ01227793 GDIQ01225644 GDIQ01210290 GDIQ01191691 JAI70394.1 JAK41435.1 GDIQ01245981 GDIQ01227794 GDIQ01210291 GDIQ01207807 GDIQ01206458 GDIQ01203248 GDIQ01191690 GDIQ01132238 GDIQ01113476 GDIP01089236 JAK41434.1 JAM14479.1 GDIP01231880 JAI91521.1 GDIP01030205 JAM73510.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JBI3
A0A076JVB8
D1MBK3
A0A3S2P1P8
S4PK99
Q86G85
+ More
A2TIK8 C6KE04 D2DMM8 A0A2A4K6E0 A0A023UG44 A0A1E1WNR2 A0A2H1WGL4 A0A194PEH3 A0A194RGI2 F8TAD6 A0A1W4XHH0 A0A1W4XS08 D6X0D6 V5G6J5 V5GVJ5 A0A1B6ET25 A0A1B6FVP7 A0A1B6JXH3 A0A1B0AX62 A0A154PMC2 A0A2P8Y2E2 E2A6Z5 A0A2P9A990 A0A336MWD6 B4NEJ2 A0A1J1HRB8 A0A1B6MUI8 A0A0P5CDN4 A0A0N8A8C6 E9GI17 A0A336MBS4 A0A336M8L1 A0A1I8PFU3 A0A0P6IR12 A0A164YYE1 A0A0P5IU01 A0A0N8B5P6 A0A0P4Y6Q1 A0A0P6AIE1
A2TIK8 C6KE04 D2DMM8 A0A2A4K6E0 A0A023UG44 A0A1E1WNR2 A0A2H1WGL4 A0A194PEH3 A0A194RGI2 F8TAD6 A0A1W4XHH0 A0A1W4XS08 D6X0D6 V5G6J5 V5GVJ5 A0A1B6ET25 A0A1B6FVP7 A0A1B6JXH3 A0A1B0AX62 A0A154PMC2 A0A2P8Y2E2 E2A6Z5 A0A2P9A990 A0A336MWD6 B4NEJ2 A0A1J1HRB8 A0A1B6MUI8 A0A0P5CDN4 A0A0N8A8C6 E9GI17 A0A336MBS4 A0A336M8L1 A0A1I8PFU3 A0A0P6IR12 A0A164YYE1 A0A0P5IU01 A0A0N8B5P6 A0A0P4Y6Q1 A0A0P6AIE1
Ontologies
GO
GO:0007229
GO:0007155
GO:0016021
GO:0009986
GO:0033627
GO:0007160
GO:0005925
GO:0005178
GO:0008305
GO:0016477
GO:0007298
GO:0021551
GO:0007015
GO:0016328
GO:0031252
GO:0035160
GO:0007508
GO:0003344
GO:1990430
GO:0007431
GO:0007377
GO:0016203
GO:0035099
GO:0006930
GO:0090129
GO:0007411
GO:0030718
GO:0005927
GO:0009925
GO:0007608
GO:0007419
GO:0051492
GO:0046982
GO:0008340
GO:0007391
GO:0035001
GO:0008360
GO:0034446
GO:0042734
GO:0045211
GO:0043034
GO:0045214
GO:0030425
GO:0030336
GO:0048803
PANTHER
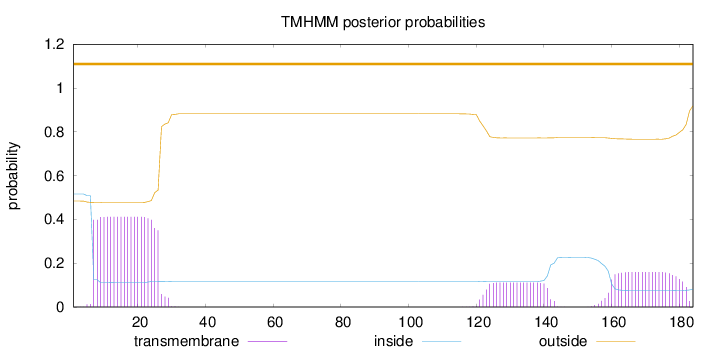
Topology
Subcellular location
Membrane
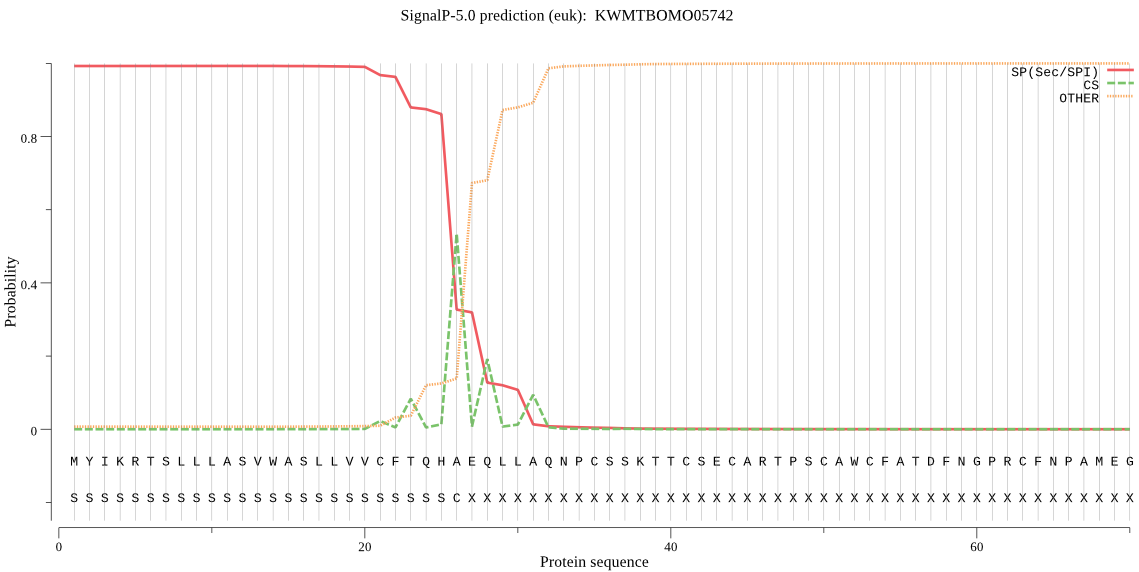
SignalP
Position: 1 - 26,
Likelihood: 0.993083
Length:
184
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.07417
Exp number, first 60 AAs:
8.25468
Total prob of N-in:
0.51620
outside
1 - 184
Population Genetic Test Statistics
Pi
144.577011
Theta
210.880056
Tajima's D
-0.932683
CLR
61.933449
CSRT
0.149842507874606
Interpretation
Uncertain
