Gene
KWMTBOMO05740
Pre Gene Modal
BGIBMGA006877
Annotation
PREDICTED:_sorting_nexin-29-like_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.741
Sequence
CDS
ATGAATAGTAAAATTTTAAATACTTTAGCTAATGCAATTGATCCTAAAGAAGAAATTATGAAAAGAGCGGAAACTGGAAAAGTATTACTCAATGAACTTTCAAAGTGTACACAAAATTGTCTCAACCGATTTGCTGGAAGATCTGAATTAGCTACTGAAGATGATCTCAGAATAATACAGCTGTGTGAAAAATGGGAAGCCATATTAAGACATGGAATAAGAACAAATTTGTCGAACTCAACCATACAAAGCTTGGTGACTGCAGGTTTAAATTTTACATTCAATATTGTAAATATAGGCAACTCATTATGGAGCTATACATGTTTGCATTTGACAAAACATGAAAAAGAAAGATTTAAAATATTGTCAAACATAAACACACCCCTTGGATATTTTAGAGCATTCTTAAGGGCTTCCTTAAATGAAAGATCATTTGAAAGATATCTGCAAAGCTGGGTGGCGCATGGCTTATTAATGGAATATTATGAAGAAGGGGCTCTTGTGAGAGATTCTGAAGCTTCCTTGAAACTTCCTAATATTGCATCTGGCTTATCATCCATTCTATTTGCGTTATCAATTGACAGACCTGAGTTGAATGAATCACAACAGTCAAGTCAAGTAGATAAAGTAGAGCATGTTATACCACTACCGACACCAGTCAAGTCCTCTGGGAACTTAAAACGGAAACCATTACGTCAAGTTATATCATTTGATAAAAATGAACGGAAGACTCAGAACATTGATCATAGTAAAGAGATACACTCAATCAACAGCAAAGATGATGCAATGTGGAACAGTGCACCGGCAACTTGCCTTAATTCTCCTGATCCTAAAATTGTGCCTCAAATATCATCAACAAGTGAACCTGCAAGCTCAGATTTCAAAAGTGGTTTCAGACATTTCTTTCCCGATAGTGTAAAAGGTATAGAAAGTCCTTCACAAATTTTAACAAAACTTTCTGAATGTGCCAAAGAAATATTTTCATCTTCAAACAGTATAGAAACAAACAATACTGCTAAAGATGATGCGTCAGAATTGAGTGTTAACAATTTAAAAATATATGAAAGTGACAGTGAGGAAGTAGCTGGTAGTATTGATGGAAGCACATCGTGCTTGGAGCTTTGCTTCACTGAAGATGAAAGTGCACCAGACGATAAATCATTACTTTCCTTGTTGGAAAATAAAGATAAACAAATTTTGATGTTACAAATGAAATGTAGCCAGTTAGAAGTAGCAAGCAAAGATAAATTTCGAAAACTTGAAAAAGTAATAATAGATCTCACCAAAGAAAACGACACAATAAAGGAGCAGTTACGACATTACATGTCCGCTGTGGATATGGGCAAAGCTCTAAGGAATAATAATGTTCGGGAGGAAGCAAATAATGACAACACTGATGAAATAAACCAGTATGAAAGAAAACTTGTTCAGGTAGCCGAGATGCATGCAGAGCTGATGGAGTTTAACCAGCATCTGGTGCAGAGGCTACAAGAATTGGAAAGCACATGTGCCCTCGAACTATTAGATTACCCTGAGTCTAATGTTAAGGCTCATATTCCCAGTGCATTTTTAGTTGGAAAGAGAACACACTCCTTTCATGTTTATCAAATTTTTCTAAAAGTTGGCAATGAAGAATGGAGCGTCTACCATAGATATGCGAAGTTCCACGAACTTCATACGCAGTTAAAAAAATGTCATCCCGATATAGCAACATACAAATTTCCACCTAAAAAAACCTTAAGAAAACGGGACGCCCATCTAGTCGAGCAGCGCCGCCTAGCGCTCCAGGCGTACCTGCGGCACGTCCTTCTCGTGCTGCCCGAGCTCAGGGAGTGCACCAGCCGGCCGCAGCTCGTCACGCTACTGCCTTTCTTTGGCACCTCCTCGACTTCCAAAGAAAATGGTGTTCACGACGTCCTCGATCGACAACACTCAAGTGACTCTGCCTCATCATACAATGTACTTTGA
Protein
MNSKILNTLANAIDPKEEIMKRAETGKVLLNELSKCTQNCLNRFAGRSELATEDDLRIIQLCEKWEAILRHGIRTNLSNSTIQSLVTAGLNFTFNIVNIGNSLWSYTCLHLTKHEKERFKILSNINTPLGYFRAFLRASLNERSFERYLQSWVAHGLLMEYYEEGALVRDSEASLKLPNIASGLSSILFALSIDRPELNESQQSSQVDKVEHVIPLPTPVKSSGNLKRKPLRQVISFDKNERKTQNIDHSKEIHSINSKDDAMWNSAPATCLNSPDPKIVPQISSTSEPASSDFKSGFRHFFPDSVKGIESPSQILTKLSECAKEIFSSSNSIETNNTAKDDASELSVNNLKIYESDSEEVAGSIDGSTSCLELCFTEDESAPDDKSLLSLLENKDKQILMLQMKCSQLEVASKDKFRKLEKVIIDLTKENDTIKEQLRHYMSAVDMGKALRNNNVREEANNDNTDEINQYERKLVQVAEMHAELMEFNQHLVQRLQELESTCALELLDYPESNVKAHIPSAFLVGKRTHSFHVYQIFLKVGNEEWSVYHRYAKFHELHTQLKKCHPDIATYKFPPKKTLRKRDAHLVEQRRLALQAYLRHVLLVLPELRECTSRPQLVTLLPFFGTSSTSKENGVHDVLDRQHSSDSASSYNVL
Summary
Uniprot
A0A2H1WGY5
A0A2A4K4S4
A0A1E1W1B1
A0A1E1WRU0
A0A2A4K6G0
A0A194PDK7
+ More
A0A3S2LBM4 A0A212EPQ6 A0A194RC46 A0A0L7LQS0 A0A1W4X2G1 A0A1W4X278 A0A1Y1JX67 A0A1Y1JSY8 E2BNV2 A0A0N0BH99 E0VG85 U4UJD0 A0A2A3EIU4 A0A087ZQ69 A0A154P3Z8 A0A2C9K6S7 A0A0L7R4I4 N6UA22 T1IGL6 A0A1B6EX13 E9J8Q4 A0A151IH67 E2ACM0 A0A0A9ZI25 K7IRM4 A0A151IT09 A0A026WU31 A0A3L8DFY6 F4WAU7 A0A151XFY9 A0A2L2Y9I0 A0A1B6CB94 A0A195F0M2 A0A158P413 X1WJ48 A0A2H8U1Z0 A0A1B6HMV0 A0A224XH26 A0A0K8S6G7 A0A0K8S7Q6 A0A2S2QRD5 K1R182 A0A0P6E022 A0A0J7L894 A0A195BYZ6 A0A0K2UYL9
A0A3S2LBM4 A0A212EPQ6 A0A194RC46 A0A0L7LQS0 A0A1W4X2G1 A0A1W4X278 A0A1Y1JX67 A0A1Y1JSY8 E2BNV2 A0A0N0BH99 E0VG85 U4UJD0 A0A2A3EIU4 A0A087ZQ69 A0A154P3Z8 A0A2C9K6S7 A0A0L7R4I4 N6UA22 T1IGL6 A0A1B6EX13 E9J8Q4 A0A151IH67 E2ACM0 A0A0A9ZI25 K7IRM4 A0A151IT09 A0A026WU31 A0A3L8DFY6 F4WAU7 A0A151XFY9 A0A2L2Y9I0 A0A1B6CB94 A0A195F0M2 A0A158P413 X1WJ48 A0A2H8U1Z0 A0A1B6HMV0 A0A224XH26 A0A0K8S6G7 A0A0K8S7Q6 A0A2S2QRD5 K1R182 A0A0P6E022 A0A0J7L894 A0A195BYZ6 A0A0K2UYL9
Pubmed
EMBL
ODYU01008525
SOQ52206.1
NWSH01000130
PCG79251.1
GDQN01010298
JAT80756.1
+ More
GDQN01001346 JAT89708.1 PCG79250.1 KQ459606 KPI91431.1 RSAL01000053 RVE50103.1 AGBW02013402 OWR43482.1 KQ460398 KPJ15044.1 JTDY01000312 KOB77749.1 GEZM01101656 JAV52400.1 GEZM01101657 JAV52399.1 GL449507 EFN82635.1 KQ435756 KOX75912.1 DS235132 EEB12391.1 KB631008 KB632204 ERL83999.1 ERL90090.1 KZ288229 PBC31723.1 KQ434809 KZC06601.1 KQ414658 KOC65666.1 APGK01043444 APGK01043445 KB741014 ENN75472.1 ACPB03012364 GECZ01027369 JAS42400.1 GL769036 EFZ10794.1 KQ977636 KYN01154.1 GL438568 EFN68764.1 GBHO01001289 GBHO01001287 JAG42315.1 JAG42317.1 KQ981039 KYN10144.1 KK107109 EZA59131.1 QOIP01000008 RLU19370.1 GL888053 EGI68703.1 KQ982174 KYQ59312.1 IAAA01013464 LAA03815.1 GEDC01026584 JAS10714.1 KQ981880 KYN33911.1 ADTU01008194 ADTU01008195 ADTU01008196 ABLF02039442 GFXV01007683 MBW19488.1 GECU01031684 JAS76022.1 GFTR01007308 JAW09118.1 GBRD01017068 JAG48759.1 GBRD01017067 JAG48760.1 GGMS01011133 MBY80336.1 JH817156 EKC37234.1 GDIQ01070107 JAN24630.1 LBMM01000230 KMR03603.1 KQ976394 KYM93146.1 HACA01025982 CDW43343.1
GDQN01001346 JAT89708.1 PCG79250.1 KQ459606 KPI91431.1 RSAL01000053 RVE50103.1 AGBW02013402 OWR43482.1 KQ460398 KPJ15044.1 JTDY01000312 KOB77749.1 GEZM01101656 JAV52400.1 GEZM01101657 JAV52399.1 GL449507 EFN82635.1 KQ435756 KOX75912.1 DS235132 EEB12391.1 KB631008 KB632204 ERL83999.1 ERL90090.1 KZ288229 PBC31723.1 KQ434809 KZC06601.1 KQ414658 KOC65666.1 APGK01043444 APGK01043445 KB741014 ENN75472.1 ACPB03012364 GECZ01027369 JAS42400.1 GL769036 EFZ10794.1 KQ977636 KYN01154.1 GL438568 EFN68764.1 GBHO01001289 GBHO01001287 JAG42315.1 JAG42317.1 KQ981039 KYN10144.1 KK107109 EZA59131.1 QOIP01000008 RLU19370.1 GL888053 EGI68703.1 KQ982174 KYQ59312.1 IAAA01013464 LAA03815.1 GEDC01026584 JAS10714.1 KQ981880 KYN33911.1 ADTU01008194 ADTU01008195 ADTU01008196 ABLF02039442 GFXV01007683 MBW19488.1 GECU01031684 JAS76022.1 GFTR01007308 JAW09118.1 GBRD01017068 JAG48759.1 GBRD01017067 JAG48760.1 GGMS01011133 MBY80336.1 JH817156 EKC37234.1 GDIQ01070107 JAN24630.1 LBMM01000230 KMR03603.1 KQ976394 KYM93146.1 HACA01025982 CDW43343.1
Proteomes
UP000218220
UP000053268
UP000283053
UP000007151
UP000053240
UP000037510
+ More
UP000192223 UP000008237 UP000053105 UP000009046 UP000030742 UP000242457 UP000005203 UP000076502 UP000076420 UP000053825 UP000019118 UP000015103 UP000078542 UP000000311 UP000002358 UP000078492 UP000053097 UP000279307 UP000007755 UP000075809 UP000078541 UP000005205 UP000007819 UP000005408 UP000036403 UP000078540
UP000192223 UP000008237 UP000053105 UP000009046 UP000030742 UP000242457 UP000005203 UP000076502 UP000076420 UP000053825 UP000019118 UP000015103 UP000078542 UP000000311 UP000002358 UP000078492 UP000053097 UP000279307 UP000007755 UP000075809 UP000078541 UP000005205 UP000007819 UP000005408 UP000036403 UP000078540
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H1WGY5
A0A2A4K4S4
A0A1E1W1B1
A0A1E1WRU0
A0A2A4K6G0
A0A194PDK7
+ More
A0A3S2LBM4 A0A212EPQ6 A0A194RC46 A0A0L7LQS0 A0A1W4X2G1 A0A1W4X278 A0A1Y1JX67 A0A1Y1JSY8 E2BNV2 A0A0N0BH99 E0VG85 U4UJD0 A0A2A3EIU4 A0A087ZQ69 A0A154P3Z8 A0A2C9K6S7 A0A0L7R4I4 N6UA22 T1IGL6 A0A1B6EX13 E9J8Q4 A0A151IH67 E2ACM0 A0A0A9ZI25 K7IRM4 A0A151IT09 A0A026WU31 A0A3L8DFY6 F4WAU7 A0A151XFY9 A0A2L2Y9I0 A0A1B6CB94 A0A195F0M2 A0A158P413 X1WJ48 A0A2H8U1Z0 A0A1B6HMV0 A0A224XH26 A0A0K8S6G7 A0A0K8S7Q6 A0A2S2QRD5 K1R182 A0A0P6E022 A0A0J7L894 A0A195BYZ6 A0A0K2UYL9
A0A3S2LBM4 A0A212EPQ6 A0A194RC46 A0A0L7LQS0 A0A1W4X2G1 A0A1W4X278 A0A1Y1JX67 A0A1Y1JSY8 E2BNV2 A0A0N0BH99 E0VG85 U4UJD0 A0A2A3EIU4 A0A087ZQ69 A0A154P3Z8 A0A2C9K6S7 A0A0L7R4I4 N6UA22 T1IGL6 A0A1B6EX13 E9J8Q4 A0A151IH67 E2ACM0 A0A0A9ZI25 K7IRM4 A0A151IT09 A0A026WU31 A0A3L8DFY6 F4WAU7 A0A151XFY9 A0A2L2Y9I0 A0A1B6CB94 A0A195F0M2 A0A158P413 X1WJ48 A0A2H8U1Z0 A0A1B6HMV0 A0A224XH26 A0A0K8S6G7 A0A0K8S7Q6 A0A2S2QRD5 K1R182 A0A0P6E022 A0A0J7L894 A0A195BYZ6 A0A0K2UYL9
PDB
6EE0
E-value=7.99219e-11,
Score=164
Ontologies
GO
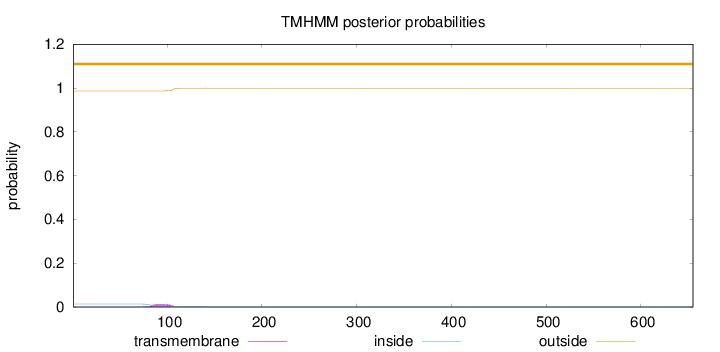
Topology
Length:
655
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29809
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01349
outside
1 - 655
Population Genetic Test Statistics
Pi
182.374968
Theta
166.019649
Tajima's D
-0.654146
CLR
117.219684
CSRT
0.207639618019099
Interpretation
Uncertain
