Gene
KWMTBOMO05734
Pre Gene Modal
BGIBMGA006812
Annotation
PREDICTED:_allantoinase_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.618
Sequence
CDS
ATGGTGTTAAGCCGACAACTGTTTTTGAGTAAGCGTCTAGTTGTCGAAGACAAGGTTTTCGATGGCGGTATTTTAGTCAATAGCTATGGTGTTATCGAAAGAGTTCTTGAAAGAAAAGAAGTTAGTCGACTTTTGGACGAAAGTGGTGAAAATATTAAGGTAGTGGACGGTGGTGATTTAGCGCTGATAGCGGGCATCGTTGATTCGCACGTCCATATTAACGAGCCGGGTCGAACCGCTTGGGAGGGATTCCAAACGGCAACTAAGGCTGCAGCTGCTGGTGGGATTACCACGATAATCGATATGCCCTTGAATTCGATACCGCCAACTACCTCGTTGGAAAACTTGAAAATTAAAGCAACAGCCGCTAAAGGAAATGTATTTGTTGATGTAGGATTCTGGGGAGGAGTGATACCGCAAAATCATTTCCATGCTGAAATCGACTGCGCTTACGATCAAGGCCATAAAGAAAAAATGAGAAAGAAAGATCTTGAAGAATATCACACATTCCTGGAATCGAGACCTGCAAGCATGGAACTCGAAGCTTTCAATTTAATAAAAAAATTTGTTAAGAAACACAATGTACGCGTTCATATAGTACACGTATCATCTGCAGAAATAATCCCTTTACTTCATGCCTTGCGTCGAGCAAGAAGTGAGAACAATGGATTTTGGCGGAACGGAATTACAGCTGAAACTTGCCATCACTATTTAACATTTAGCGCTGAGCAAATACCAAAAGGGCATAGCGAATTTAAATGCGCTCCACCCATTAGAGATAAAAATAACCAGGAAAAATTATGGGATTTCCTGTTAGAAGATAAAATAGACTTAATTGTATCCGATCACTCCCCGTGTACTCCAGACCTTAAGTGCAACAAAAATCTTATGGAAGCGTGGGGTGGAATTTCTTCCGTGCAATTCGGGCTATCATTGTTTTGGACAGCAGCCGAAGCTCGTGGCTTGGATTTGACTTCGATCAGTAAATACTTAAGTTCTGGACCAGCCCATTTGTGTAGTATACAACACAAGAAGGGTGCTATTAAGCCTGGTCTAGATGCCGATCTTGTGTTTTTCGATCCAGAGGCTGAATTTGTCATTACTACTGACATTATCCGGCACAAAAATAAGCTAACACCGTATCTGAATATGAAACTCAAGGGCGTAGTAAAAGAAACATATCTTCGTGGTCAATTGATATACAGAGATGGCAATGTTATTGGTCAACCTATAGGAGAATTATTACTGAAAGAATCTTAA
Protein
MVLSRQLFLSKRLVVEDKVFDGGILVNSYGVIERVLERKEVSRLLDESGENIKVVDGGDLALIAGIVDSHVHINEPGRTAWEGFQTATKAAAAGGITTIIDMPLNSIPPTTSLENLKIKATAAKGNVFVDVGFWGGVIPQNHFHAEIDCAYDQGHKEKMRKKDLEEYHTFLESRPASMELEAFNLIKKFVKKHNVRVHIVHVSSAEIIPLLHALRRARSENNGFWRNGITAETCHHYLTFSAEQIPKGHSEFKCAPPIRDKNNQEKLWDFLLEDKIDLIVSDHSPCTPDLKCNKNLMEAWGGISSVQFGLSLFWTAAEARGLDLTSISKYLSSGPAHLCSIQHKKGAIKPGLDADLVFFDPEAEFVITTDIIRHKNKLTPYLNMKLKGVVKETYLRGQLIYRDGNVIGQPIGELLLKES
Summary
Uniprot
H9JBB7
A0A3S2NMG2
A0A2A4K6D3
A0A212EN67
A0A194PEI3
A0A2H1VYM4
+ More
A0A194RB47 S4P2A0 A0A194RC55 A0A194PFG3 A0A212EN58 A0A2A4K639 A0A067R3H9 A0A2J7RJ99 A0A2J7RJ97 C3YQZ7 A0A1L8DXS6 Q8I6V5 A0A1L8DXX7 K7IT29 A0A232FIH1 A0A068TK19 A0A1Y1K3C0 A0A3Q2G0D7 A0A182W598 A0A023EST7 U4KJB9 M3XKJ9 A0A182GP83 A0A182RQ70 A0A3Q3A2M6 A0A2I4BFI7 Q171F8 Q5PR45 F1RB68 A0A182MPY1 H3AEL7 H2TVH1 A0A195F4A7 A0A3P8VGB8 A0A3P8U0D8 A0A158NJX8 A0A151I862 A0A195DW63 A0A182PR28 A0A182XY17 G3NKQ6 A0A3Q1AKY4 A0A3Q3RYN2 A0A3Q3GY48 A0A3B3I556 A0A1Q3FS53 H3CC73 A0A3Q1FHG8 B0W9J6 A0A182Q5B8 A0A0S7LIB5 A0A182K9C4 A0A3B3TBG1 A0A3B3DD81 A0A3P8VLJ8 A0A3Q2QET1 A0A3B4DBS4 A0A182N0B4 A0A1S3JJ52 K4G5M6 A0A146N609 A0A3Q1JI77 A0A2D0S1K5 A0A3P9KFR3 A0A3Q0RCS1 A0A336LJE0 W5MHP2 A0A084VLM1 A0A182UV22 A0A182WTM0 G3NKP4 A0A3Q2YZQ6 A0A3P9IW08 A0A315V662 A0A151XE50 Q7QF02 A0A182TE02 M3ZNF5 G3W519 A0A182IAG3 A0A3M6UUG5 A0A3P8ZM39 A0A3B4WKH1 A0A210PXM9 A0A3Q3CQZ8 K4FYD3 A0A3Q3WAP7 A0A3B4GW76 A0A3P8RB74 A0A3B1K294 A0A087YL43 A0A182IVY8 A0A1B0C997 V9KKR8
A0A194RB47 S4P2A0 A0A194RC55 A0A194PFG3 A0A212EN58 A0A2A4K639 A0A067R3H9 A0A2J7RJ99 A0A2J7RJ97 C3YQZ7 A0A1L8DXS6 Q8I6V5 A0A1L8DXX7 K7IT29 A0A232FIH1 A0A068TK19 A0A1Y1K3C0 A0A3Q2G0D7 A0A182W598 A0A023EST7 U4KJB9 M3XKJ9 A0A182GP83 A0A182RQ70 A0A3Q3A2M6 A0A2I4BFI7 Q171F8 Q5PR45 F1RB68 A0A182MPY1 H3AEL7 H2TVH1 A0A195F4A7 A0A3P8VGB8 A0A3P8U0D8 A0A158NJX8 A0A151I862 A0A195DW63 A0A182PR28 A0A182XY17 G3NKQ6 A0A3Q1AKY4 A0A3Q3RYN2 A0A3Q3GY48 A0A3B3I556 A0A1Q3FS53 H3CC73 A0A3Q1FHG8 B0W9J6 A0A182Q5B8 A0A0S7LIB5 A0A182K9C4 A0A3B3TBG1 A0A3B3DD81 A0A3P8VLJ8 A0A3Q2QET1 A0A3B4DBS4 A0A182N0B4 A0A1S3JJ52 K4G5M6 A0A146N609 A0A3Q1JI77 A0A2D0S1K5 A0A3P9KFR3 A0A3Q0RCS1 A0A336LJE0 W5MHP2 A0A084VLM1 A0A182UV22 A0A182WTM0 G3NKP4 A0A3Q2YZQ6 A0A3P9IW08 A0A315V662 A0A151XE50 Q7QF02 A0A182TE02 M3ZNF5 G3W519 A0A182IAG3 A0A3M6UUG5 A0A3P8ZM39 A0A3B4WKH1 A0A210PXM9 A0A3Q3CQZ8 K4FYD3 A0A3Q3WAP7 A0A3B4GW76 A0A3P8RB74 A0A3B1K294 A0A087YL43 A0A182IVY8 A0A1B0C997 V9KKR8
Pubmed
19121390
22118469
26354079
23622113
24845553
18563158
+ More
14871617 20075255 28648823 28004739 24945155 9215903 26483478 17510324 18182492 23594743 21551351 24487278 21347285 25244985 17554307 15496914 29240929 29451363 23056606 24438588 29703783 12364791 14747013 17210077 23542700 21709235 30382153 25069045 28812685 25329095 24402279
14871617 20075255 28648823 28004739 24945155 9215903 26483478 17510324 18182492 23594743 21551351 24487278 21347285 25244985 17554307 15496914 29240929 29451363 23056606 24438588 29703783 12364791 14747013 17210077 23542700 21709235 30382153 25069045 28812685 25329095 24402279
EMBL
BABH01030921
BABH01030922
RSAL01000254
RVE43443.1
NWSH01000130
PCG79232.1
+ More
AGBW02013719 OWR42919.1 KQ459606 KPI91438.1 ODYU01005260 SOQ45935.1 KQ460398 KPJ15053.1 GAIX01012125 JAA80435.1 KPJ15054.1 KPI91439.1 OWR42918.1 PCG79233.1 KK853018 KDR12419.1 NEVH01002994 PNF40904.1 PNF40903.1 GG666545 EEN57239.1 GFDF01002817 JAV11267.1 AY151136 AAN72834.1 GFDF01002818 JAV11266.1 AAZX01003655 NNAY01000179 OXU30258.1 HG965799 CDO39395.1 GEZM01094080 JAV55949.1 GAPW01001251 JAC12347.1 HF678436 CCT61359.1 AFYH01081025 AFYH01081026 JXUM01077892 JXUM01077893 KQ563032 KXJ74606.1 EF676029 CH477456 ABV44609.1 EAT40633.1 BC086832 BC165735 AAH86832.1 AAI65735.1 CU467043 AXCM01000049 KQ981820 KYN35298.1 ADTU01018461 ADTU01018462 ADTU01018463 ADTU01018464 ADTU01018465 KQ978390 KYM94313.1 KQ980304 KYN16799.1 GFDL01004782 JAV30263.1 DS231864 EDS40221.1 AXCN02001229 GBYX01167673 JAO89216.1 JX209380 AFM87694.1 GCES01159440 JAQ26882.1 UFQT01000015 SSX17725.1 AHAT01015231 ATLV01014525 KE524974 KFB38865.1 NHOQ01002838 PWA14419.1 KQ982254 KYQ58637.1 AAAB01008846 EAA06409.5 AEFK01136930 AEFK01136931 APCN01001235 RCHS01000689 RMX57239.1 NEDP02005415 OWF41243.1 JX052994 AFK11222.1 AYCK01022118 AJWK01002077 JW866126 AFO98643.1
AGBW02013719 OWR42919.1 KQ459606 KPI91438.1 ODYU01005260 SOQ45935.1 KQ460398 KPJ15053.1 GAIX01012125 JAA80435.1 KPJ15054.1 KPI91439.1 OWR42918.1 PCG79233.1 KK853018 KDR12419.1 NEVH01002994 PNF40904.1 PNF40903.1 GG666545 EEN57239.1 GFDF01002817 JAV11267.1 AY151136 AAN72834.1 GFDF01002818 JAV11266.1 AAZX01003655 NNAY01000179 OXU30258.1 HG965799 CDO39395.1 GEZM01094080 JAV55949.1 GAPW01001251 JAC12347.1 HF678436 CCT61359.1 AFYH01081025 AFYH01081026 JXUM01077892 JXUM01077893 KQ563032 KXJ74606.1 EF676029 CH477456 ABV44609.1 EAT40633.1 BC086832 BC165735 AAH86832.1 AAI65735.1 CU467043 AXCM01000049 KQ981820 KYN35298.1 ADTU01018461 ADTU01018462 ADTU01018463 ADTU01018464 ADTU01018465 KQ978390 KYM94313.1 KQ980304 KYN16799.1 GFDL01004782 JAV30263.1 DS231864 EDS40221.1 AXCN02001229 GBYX01167673 JAO89216.1 JX209380 AFM87694.1 GCES01159440 JAQ26882.1 UFQT01000015 SSX17725.1 AHAT01015231 ATLV01014525 KE524974 KFB38865.1 NHOQ01002838 PWA14419.1 KQ982254 KYQ58637.1 AAAB01008846 EAA06409.5 AEFK01136930 AEFK01136931 APCN01001235 RCHS01000689 RMX57239.1 NEDP02005415 OWF41243.1 JX052994 AFK11222.1 AYCK01022118 AJWK01002077 JW866126 AFO98643.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053268
UP000053240
+ More
UP000027135 UP000235965 UP000001554 UP000002358 UP000215335 UP000265020 UP000075920 UP000008672 UP000069940 UP000249989 UP000075900 UP000264800 UP000192220 UP000008820 UP000000437 UP000075883 UP000005226 UP000078541 UP000265120 UP000265080 UP000005205 UP000078542 UP000078492 UP000075885 UP000076408 UP000007635 UP000257160 UP000261640 UP000261660 UP000001038 UP000007303 UP000257200 UP000002320 UP000075886 UP000075881 UP000261540 UP000261560 UP000265000 UP000261440 UP000075884 UP000085678 UP000265040 UP000221080 UP000265180 UP000261340 UP000018468 UP000030765 UP000075903 UP000076407 UP000264820 UP000265200 UP000075809 UP000007062 UP000075902 UP000002852 UP000007648 UP000075840 UP000275408 UP000265140 UP000261360 UP000242188 UP000264840 UP000261620 UP000261460 UP000265100 UP000018467 UP000028760 UP000075880 UP000092461
UP000027135 UP000235965 UP000001554 UP000002358 UP000215335 UP000265020 UP000075920 UP000008672 UP000069940 UP000249989 UP000075900 UP000264800 UP000192220 UP000008820 UP000000437 UP000075883 UP000005226 UP000078541 UP000265120 UP000265080 UP000005205 UP000078542 UP000078492 UP000075885 UP000076408 UP000007635 UP000257160 UP000261640 UP000261660 UP000001038 UP000007303 UP000257200 UP000002320 UP000075886 UP000075881 UP000261540 UP000261560 UP000265000 UP000261440 UP000075884 UP000085678 UP000265040 UP000221080 UP000265180 UP000261340 UP000018468 UP000030765 UP000075903 UP000076407 UP000264820 UP000265200 UP000075809 UP000007062 UP000075902 UP000002852 UP000007648 UP000075840 UP000275408 UP000265140 UP000261360 UP000242188 UP000264840 UP000261620 UP000261460 UP000265100 UP000018467 UP000028760 UP000075880 UP000092461
PRIDE
Interpro
ProteinModelPortal
H9JBB7
A0A3S2NMG2
A0A2A4K6D3
A0A212EN67
A0A194PEI3
A0A2H1VYM4
+ More
A0A194RB47 S4P2A0 A0A194RC55 A0A194PFG3 A0A212EN58 A0A2A4K639 A0A067R3H9 A0A2J7RJ99 A0A2J7RJ97 C3YQZ7 A0A1L8DXS6 Q8I6V5 A0A1L8DXX7 K7IT29 A0A232FIH1 A0A068TK19 A0A1Y1K3C0 A0A3Q2G0D7 A0A182W598 A0A023EST7 U4KJB9 M3XKJ9 A0A182GP83 A0A182RQ70 A0A3Q3A2M6 A0A2I4BFI7 Q171F8 Q5PR45 F1RB68 A0A182MPY1 H3AEL7 H2TVH1 A0A195F4A7 A0A3P8VGB8 A0A3P8U0D8 A0A158NJX8 A0A151I862 A0A195DW63 A0A182PR28 A0A182XY17 G3NKQ6 A0A3Q1AKY4 A0A3Q3RYN2 A0A3Q3GY48 A0A3B3I556 A0A1Q3FS53 H3CC73 A0A3Q1FHG8 B0W9J6 A0A182Q5B8 A0A0S7LIB5 A0A182K9C4 A0A3B3TBG1 A0A3B3DD81 A0A3P8VLJ8 A0A3Q2QET1 A0A3B4DBS4 A0A182N0B4 A0A1S3JJ52 K4G5M6 A0A146N609 A0A3Q1JI77 A0A2D0S1K5 A0A3P9KFR3 A0A3Q0RCS1 A0A336LJE0 W5MHP2 A0A084VLM1 A0A182UV22 A0A182WTM0 G3NKP4 A0A3Q2YZQ6 A0A3P9IW08 A0A315V662 A0A151XE50 Q7QF02 A0A182TE02 M3ZNF5 G3W519 A0A182IAG3 A0A3M6UUG5 A0A3P8ZM39 A0A3B4WKH1 A0A210PXM9 A0A3Q3CQZ8 K4FYD3 A0A3Q3WAP7 A0A3B4GW76 A0A3P8RB74 A0A3B1K294 A0A087YL43 A0A182IVY8 A0A1B0C997 V9KKR8
A0A194RB47 S4P2A0 A0A194RC55 A0A194PFG3 A0A212EN58 A0A2A4K639 A0A067R3H9 A0A2J7RJ99 A0A2J7RJ97 C3YQZ7 A0A1L8DXS6 Q8I6V5 A0A1L8DXX7 K7IT29 A0A232FIH1 A0A068TK19 A0A1Y1K3C0 A0A3Q2G0D7 A0A182W598 A0A023EST7 U4KJB9 M3XKJ9 A0A182GP83 A0A182RQ70 A0A3Q3A2M6 A0A2I4BFI7 Q171F8 Q5PR45 F1RB68 A0A182MPY1 H3AEL7 H2TVH1 A0A195F4A7 A0A3P8VGB8 A0A3P8U0D8 A0A158NJX8 A0A151I862 A0A195DW63 A0A182PR28 A0A182XY17 G3NKQ6 A0A3Q1AKY4 A0A3Q3RYN2 A0A3Q3GY48 A0A3B3I556 A0A1Q3FS53 H3CC73 A0A3Q1FHG8 B0W9J6 A0A182Q5B8 A0A0S7LIB5 A0A182K9C4 A0A3B3TBG1 A0A3B3DD81 A0A3P8VLJ8 A0A3Q2QET1 A0A3B4DBS4 A0A182N0B4 A0A1S3JJ52 K4G5M6 A0A146N609 A0A3Q1JI77 A0A2D0S1K5 A0A3P9KFR3 A0A3Q0RCS1 A0A336LJE0 W5MHP2 A0A084VLM1 A0A182UV22 A0A182WTM0 G3NKP4 A0A3Q2YZQ6 A0A3P9IW08 A0A315V662 A0A151XE50 Q7QF02 A0A182TE02 M3ZNF5 G3W519 A0A182IAG3 A0A3M6UUG5 A0A3P8ZM39 A0A3B4WKH1 A0A210PXM9 A0A3Q3CQZ8 K4FYD3 A0A3Q3WAP7 A0A3B4GW76 A0A3P8RB74 A0A3B1K294 A0A087YL43 A0A182IVY8 A0A1B0C997 V9KKR8
PDB
3E74
E-value=2.33662e-56,
Score=554
Ontologies
PATHWAY
GO
PANTHER
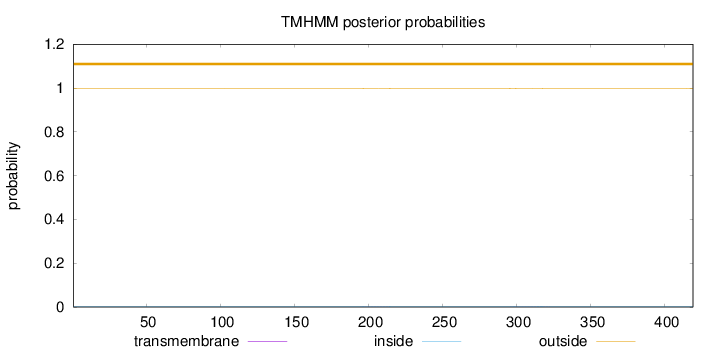
Topology
Length:
419
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03402
Exp number, first 60 AAs:
0.00094
Total prob of N-in:
0.00170
outside
1 - 419
Population Genetic Test Statistics
Pi
173.463881
Theta
154.000965
Tajima's D
0.495309
CLR
0.44693
CSRT
0.508224588770561
Interpretation
Uncertain
