Pre Gene Modal
BGIBMGA006813
Annotation
Nuclear_pore_complex_protein_Nup155_[Papilio_xuthus]
Full name
Nuclear pore complex protein Nup154
+ More
Nuclear pore complex protein Nup155
Nuclear pore complex protein Nup155
Alternative Name
154 kDa nucleoporin
Nucleoporin Nup154
Tulipan
155 kDa nucleoporin
Nucleoporin Nup155
Nucleoporin Nup154
Tulipan
155 kDa nucleoporin
Nucleoporin Nup155
Location in the cell
Nuclear Reliability : 1.442 PlasmaMembrane Reliability : 1.462
Sequence
CDS
ATGCAGCTAGAATCTTTGGAGCTGGCTGGCTGTACGCTTGACCGGTACATTAGCTTTGATAATTCGCGCCCTTCCTTTTTGGAGTTAACTGGCATTGCAGCACAACGGAGTCCAACATGCAGTGGGTTAAATGCTGGCGATTACCGCAACCTTGCTGCTCTCCTGAACCATCCTAACTTGTCCCAGTTGAAAATTCTCAACAAAGTGCCTCTGCCACCAGAGATTATGGAACACTTTGCTCATATGCAATGCCATTGTTTAATGGGAGTTTTCCCTGAAATTAGCAGAGTTTGGCTTGCAATAGACAGCAATATATATGTTTGGGCATTTGAACATGGGAGCGATGTTGCATATTTCGATGGATTAGGAGAAACCATAGTCAGTGTTGGCTTAGTTAAACCAAAACCAGGGGTGTTTCAGAATTTTGTCAAGTATTTGCTTGTTCTCACGACAACAGTGGAAATTGTTGTATTAGGTGTGACATTTTCATCAAACAAAAGCGATTCTCCAAGAGAATTTCAAGAAATACATCTTGTCTCAGAACCTGTCTTTGTTCTGCCAACTGATGGTGTGTCAATGATGTGTGTTAAATCCACTTCAAGAGGTCGAATATTTATGGGAGGTAAAGATGGCTGTCTGTATGAAATAACCTATCAGGCACAATTAGGATGGTTTGGTAAACATTGCAAGAAAGTGAACCATTCCACCAGCACTCTTTCATTCCTAGTACCATCATTTCTAAATGCCGCATTATATGACGAGGATCCGATAGTAAAAATTGAAGTTGATGATTCCCGGAACATTTTGTATACACTCAGTGAAAAAGGCTGTATAGAAGTATTTGATCTTGGCAAGGATGGTGACAGCCTCAGTCGAGTTGTAAGGCTAACACAAGGAAAGATTGTATCTGCAGCTGTGGATATAGTGAAAACTTTGGAAGTAAGCAACTTCAAACCTGTCATAGCAATTTCTGCTGTAGATGAATCAGAGTCTGACCATTTGAATTTAGTAGCAGTCACACAGACTGGAGCAAGATTATATTTTAGCACGGGAAGTGGAGATACAAACCAAAGTGGTATACAGAGACCACAGTACTTGACAATGCTGCATGTAAGGCTGCCCCCTGGATTCACTCCTAATTCATCAGTGTTAAAGCCAAAACAAGTTCACAGTGCTGTTTACGAAAATGGTTCCCTGGTAATGATATGTTCATCCGGCGGCGGCGAGGCGGAGACTCTGTGGTGCTTGTCTCGCGTCATCCCGGGCAGCGCGGCCTTCAGCGAGGCGCACACCGTGCTGGCGCTGGACGGGCCCGCCTGGGCGCTTACTGCGCTGCCTCACCAGCATGCCGCTCACATATCCAACGCTCTTCTCACTAAAAAAGAAGTGTGGTGTGTCTCGCGATGGGCAGTAGTGACAGGGGCGGGAGCCTGTATCATAACAGCCGGAGCTGCTCCAGACATACTACGAACATTACTTAGAGACTGTCGGGGTCCTGAAGCACATGCTGTCAAGGACTTCTTTCAGCTGCACAGTGTAGAACAAGCATGCGCATGCGCCTTGTATTTGGCGTGCGAGGATACCACCAGCAGTGACCTCTCAATCAGCGAGTGGGCTACCAGAGCATTCTTCTTATATGGGACCCAGATGACTCCTGCTCCAATCTATCAGCACTCTTCACAGATGCCTTCCAGTATAGGTTCACCCAAATTTTCACCGCGACAAATGTCAACGCCCATGTCGAGAGGACCGCAGGGTCAAATTCAGGGACCGCAAGCAAACAAATTCAATCAGTCGTACCAACAACACATGAACCAGTCATTCCCGGGGTCGCCGAACCACTCGATGATGACGCAACAGCCGCAATACTCTATGAGTCCGACCCCGTACAACCAGACGTCGTTCAACGGAAACATCACTCAGCAACAAAGAGACCCTAACTACCCTGTGCCAATTGAATTTAGTGCGAAACACAACGGCCTCTATATTTACGTCGGTCGAATACTGTTGCCGATATGGAATTTAAAATGCGTCTCGAAATCCCAGACTCCGGACAGTAAAGAATTCATGTCTAGTCAAGTGACAGGCGAGGACTGTGCGTACATAGCCAAGAAATTGCAACGTGTGGCGGCGTTCATGCAGCGCACATTAGCGCCGCAACATTCTGAAGAGTACACATCGCTAAATGCATTGAAGTTATTTGTCTCAATGGCAATAGAAATGTTGAATCTATGGAGAGTACTATGTGAGCACCAGTTTCACGTGATAGCCGCGAGCCTGCCCGCAGATCAGCAGAATGCTCTTCAAGCAGCAAGTTTTAGAGAATTACTGGTTGGCGGTCAGGAAGTAGCATGCTTACTTCTAGGATCTTTGGTGGCAGGATATCTGCGTGACAATGCTTCTGTCGATGGAATTTCACAGAAACTACGACAATTATGTCCAACATTATACAGACAAGAAGACGCTACTTGCTCCAAGGCCAACGAACTCTTGATGTTTGCCAAACAACAAAAGAATCCCACGGAAAGGGAAGAAATGTTGCAGCATGCCCTAAAACTGTGTAAAGATGTGGCCCCAAATGTGAACCTGCCATTGGTTTGTTCGAAACTGGCATCTGTTGGGTTTTACTCGGGGGTGGTGGAACTGTGCGCTACTTGTGCCTGCAAACTCGATCCTCAGGATAAAGCACTGCATTACTATAAAAGTGATCAGCCCACGCAAGATCGAGAAGGACATCTAGCTTATTACAGAAGAATGGAAATTTATCGCGAAGTGTGTGCTGCTCTGGAGCGTCTTTACGAGCGCAGCAACGAAGCGCCGGGCGCCGAGCCCTCCCCGCACCGAGCGCCCTCGCAGCACTCCGCCGACGCTGACGTCACTCTGTCCCCAGCTGATGCCAACTATCAAGGTAGAAAGCTGGTGTGGGAGTGCTTGTCACGAGATGACGAACTGCTGCATGTCGCCGTTTATGAATGGCTAGTCTCCAAGAATCTCGGTTCGGAATTATTGTCGCTGGGGAGGGCGCCGCCTGCGTCATTGCGGACGTACCTGCAGGCGGCGGCTAGATCGGCGCCGCCCGCAGCTTCCCTCCACCTACTGGACCTGCTGTGGAAGGTCTTAGAGAAGTGCGGAGATCATCTCGCAGCTGCACAAGTCCTAGAGGGACTCGCTACTAGACCGGGGTCGGGCGCGACGCTGGCGCAGCGCATGTCGTGGCTGTGCGCGGGCGTGGTGTGCGTGCGCGGCGCGGGCGCGGGCGGCGGGGAGGTGGTGCGGCGGCTGGAGGAGGCGGGCGAGGTGGCGCGCGTGCAGCGGGCCGTGCGCGCCGCCGCCGCCGCCCTGCCCGCCGCGCGCCGCCCCCCGCCGCCGCACATGCAGCGCCTGGACGACGAGCTGCTCGAGATCACGCAGCTGTATGAGGAGTACGCGGACCGCTACAACCTGTGGGAGTGCAAGCTGGCCATCGTGCAGTGCTCGGGGCACAACGACGCGCTGCTGGTCGAGAACATCTGGAGCAACATTCTGGCGGAGGCGGAGGCGGCCGCCAGCGCCCTGCCCGCGCCCGACGAGAGGCTGGCCTCGGTGCTCAGTAAAATCACCACTCTGGGACGCGAATATGTCAACACCGGACATTGTTTCCCCTTGTATTTCATTGCACGCCAGTTAGAAATAATGAGTTGTAAACTTCGAGCGGATCAGAGCATGGTGTTCAAAGCGATTCTAAGTATCGGAGTCTCGCTTGAACAAGTACTGGATATTTACATCAAGCTGGTGAGCGTGAACGAGCGCGTGTGGCTGGGCTGCGGCGAGGAGACGCACGCGTGCGGCGCGGCGGCGCGGCTGCTGGAGGCGGCGCGCGCGCACGTGCAGCCGCTCGCAGCGCACGTGCGGCGCCGCGCGCTCGCCCGCTGCAAGGACCTACACGAAGCCGCCCTGTCCGCGCTTCAGGCTCGTCCCAACACACAACACCTGATAGAAAGGTTGTCAGTCGCACAGGCGCACCTAGATCGTATGGATTAA
Protein
MQLESLELAGCTLDRYISFDNSRPSFLELTGIAAQRSPTCSGLNAGDYRNLAALLNHPNLSQLKILNKVPLPPEIMEHFAHMQCHCLMGVFPEISRVWLAIDSNIYVWAFEHGSDVAYFDGLGETIVSVGLVKPKPGVFQNFVKYLLVLTTTVEIVVLGVTFSSNKSDSPREFQEIHLVSEPVFVLPTDGVSMMCVKSTSRGRIFMGGKDGCLYEITYQAQLGWFGKHCKKVNHSTSTLSFLVPSFLNAALYDEDPIVKIEVDDSRNILYTLSEKGCIEVFDLGKDGDSLSRVVRLTQGKIVSAAVDIVKTLEVSNFKPVIAISAVDESESDHLNLVAVTQTGARLYFSTGSGDTNQSGIQRPQYLTMLHVRLPPGFTPNSSVLKPKQVHSAVYENGSLVMICSSGGGEAETLWCLSRVIPGSAAFSEAHTVLALDGPAWALTALPHQHAAHISNALLTKKEVWCVSRWAVVTGAGACIITAGAAPDILRTLLRDCRGPEAHAVKDFFQLHSVEQACACALYLACEDTTSSDLSISEWATRAFFLYGTQMTPAPIYQHSSQMPSSIGSPKFSPRQMSTPMSRGPQGQIQGPQANKFNQSYQQHMNQSFPGSPNHSMMTQQPQYSMSPTPYNQTSFNGNITQQQRDPNYPVPIEFSAKHNGLYIYVGRILLPIWNLKCVSKSQTPDSKEFMSSQVTGEDCAYIAKKLQRVAAFMQRTLAPQHSEEYTSLNALKLFVSMAIEMLNLWRVLCEHQFHVIAASLPADQQNALQAASFRELLVGGQEVACLLLGSLVAGYLRDNASVDGISQKLRQLCPTLYRQEDATCSKANELLMFAKQQKNPTEREEMLQHALKLCKDVAPNVNLPLVCSKLASVGFYSGVVELCATCACKLDPQDKALHYYKSDQPTQDREGHLAYYRRMEIYREVCAALERLYERSNEAPGAEPSPHRAPSQHSADADVTLSPADANYQGRKLVWECLSRDDELLHVAVYEWLVSKNLGSELLSLGRAPPASLRTYLQAAARSAPPAASLHLLDLLWKVLEKCGDHLAAAQVLEGLATRPGSGATLAQRMSWLCAGVVCVRGAGAGGGEVVRRLEEAGEVARVQRAVRAAAAALPAARRPPPPHMQRLDDELLEITQLYEEYADRYNLWECKLAIVQCSGHNDALLVENIWSNILAEAEAAASALPAPDERLASVLSKITTLGREYVNTGHCFPLYFIARQLEIMSCKLRADQSMVFKAILSIGVSLEQVLDIYIKLVSVNERVWLGCGEETHACGAAARLLEAARAHVQPLAAHVRRRALARCKDLHEAALSALQARPNTQHLIERLSVAQAHLDRMD
Summary
Description
Component of the nuclear pore complex (PubMed:17410542). Has a role in the organization of the inner nuclear membrane proteins at the nuclear envelope (PubMed:22718353). In germ cells, plays a role in the nuclear localization of components of the dpp signaling pathways, such as Medea and phosphorylated Mad (PubMed:21696798). Binds to chromatin, and together with Nup62 and Nup93-1, contributes to karyosome morphology and chromatin organization including attachment to the nuclear envelope in oocytes and nurse cells (PubMed:22718353, PubMed:26341556). Has a role in female fertility including egg chamber development; in nurse cells, has a role in the organization of F-actin in subcortical and cytoplasmic actin filaments important for the transfer of cytoplasm from nurse cells to the growing oocytes (PubMed:9732281, PubMed:10511559, PubMed:17410542, PubMed:17277377). Has a role in male spermatogenesis and fertility (PubMed:9732281, PubMed:10511559). Has a role in germ line cell proliferation (PubMed:21696798).
Essential component of nuclear pore complex. Could be essessential for embryogenesis. Nucleoporins may be involved both in binding and translocating proteins during nucleocytoplasmic transport.
Essential component of nuclear pore complex. Could be essessential for embryogenesis. Nucleoporins may be involved both in binding and translocating proteins during nucleocytoplasmic transport.
Subunit
Interacts (via N-terminus) with Nup93-1 (PubMed:22718353). Interacts with Nup35 (PubMed:22718353). Interacts with cup (PubMed:17277377).
Interacts with GLE1 and NUP35/NUP53. Able to form a heterotrimer with GLE1 and NUPL2 in vitro (By similarity). Forms a complex with NUP35, NUP93, NUP205 and lamin B (By similarity).
Interacts with GLE1. Able to form a heterotrimer with GLE1 and NUPL2 in vitro. Forms a complex with NUP35, NUP93, NUP205 and lamin B.
Interacts with GLE1 and NUP35/NUP53. Able to form a heterotrimer with GLE1 and NUPL2 in vitro (By similarity). Forms a complex with NUP35, NUP93, NUP205 and lamin B (By similarity).
Interacts with GLE1. Able to form a heterotrimer with GLE1 and NUPL2 in vitro. Forms a complex with NUP35, NUP93, NUP205 and lamin B.
Similarity
Belongs to the non-repetitive/WGA-negative nucleoporin family.
Keywords
Chromosome
Complete proteome
Cytoplasm
Membrane
mRNA transport
Nuclear pore complex
Nucleus
Protein transport
Reference proteome
Translocation
Transport
Disulfide bond
Glycoprotein
Phosphoprotein
3D-structure
Alternative splicing
Atrial fibrillation
Disease mutation
Isopeptide bond
Ubl conjugation
Feature
chain Nuclear pore complex protein Nup154
splice variant In isoform 2.
sequence variant In ATFB15; fails to accumulate at various foci of the nuclear envelope and is diffusely distributed in the cytoplasm; shows significantly reduced permeability of the nuclear envelope compared to wild-type; dbSNP:rs587777339.
splice variant In isoform 2.
sequence variant In ATFB15; fails to accumulate at various foci of the nuclear envelope and is diffusely distributed in the cytoplasm; shows significantly reduced permeability of the nuclear envelope compared to wild-type; dbSNP:rs587777339.
Uniprot
A0A194RC55
A0A194PFG3
A0A212EN81
A0A2A4K5N8
A0A2H1VZ07
H9JBB8
+ More
A0A0C9RA96 K7IWU3 A0A232FDD8 A0A026VTC4 A0A158NNH4 A0A151I9Q5 A0A151WWN5 A0A195B3A7 A0A195DKN5 E0VPL1 A0A195F702 A0A2J7R1G1 E2AUU0 A0A1Q3EYF6 B0W0E9 A0A034WBK6 A0A336LXS5 A0A336KFW9 E2BSQ0 W4VRP7 U4U5L9 A0A0L7QX73 C3YM21 A0A2A3ETX5 A0A1S3J6G6 A0A0M9AD09 A0A1B6H098 V3ZZP8 D6WBT9 B3MJZ9 A0A1W4WA12 A0A1W4W511 A0A336MUH0 A0A1Y1MLT3 B3N4Z6 B4P155 A0A1B6CAW0 A0A0J9R0K0 Q17JY3 B4HWZ6 Q7Q3R7 A0A088A0M9 G1PPP5 N6TXK7 H2PFD5 Q9V463 A0A1W4ZT04 A0A1W4ZS22 A0A310SV63 W5PKN2 K9J6A9 A0A2J8WBA4 B4LVB1 A0A3Q1M9F3 A0A250Y7S9 A0A3Q0D4E5 L8IGZ3 A0A3Q7NND3 Q3UL43 Q6ZQ45 A0A2R8VHH1 G1SCR7 Q99P88 A0A0G2K7T6 A0A3Q7NPA4 A0A2K5CJJ5 A0A024R071 O75694 F7CDY1 G3RZ49 A0A1S3AAS7 A0A2Y9QHF8 U3DWT8 K7AP62 A0A091CYW6 A0A2K6RRI9 U3FK31 Q80X48 A0A2K5X0U2 A0A1D5QL31 A0A2K5XFG1 G7P7D4 A0A2K5RME6 G7MPR2 A0A2U3XG59 M3YTX4 G5BTT4 A0A2U4AXJ5 U6CQH1 Q7ZWL0 I3MP94 A0A3Q3A962 A0A340WH03 A0A2Y9HJX2
A0A0C9RA96 K7IWU3 A0A232FDD8 A0A026VTC4 A0A158NNH4 A0A151I9Q5 A0A151WWN5 A0A195B3A7 A0A195DKN5 E0VPL1 A0A195F702 A0A2J7R1G1 E2AUU0 A0A1Q3EYF6 B0W0E9 A0A034WBK6 A0A336LXS5 A0A336KFW9 E2BSQ0 W4VRP7 U4U5L9 A0A0L7QX73 C3YM21 A0A2A3ETX5 A0A1S3J6G6 A0A0M9AD09 A0A1B6H098 V3ZZP8 D6WBT9 B3MJZ9 A0A1W4WA12 A0A1W4W511 A0A336MUH0 A0A1Y1MLT3 B3N4Z6 B4P155 A0A1B6CAW0 A0A0J9R0K0 Q17JY3 B4HWZ6 Q7Q3R7 A0A088A0M9 G1PPP5 N6TXK7 H2PFD5 Q9V463 A0A1W4ZT04 A0A1W4ZS22 A0A310SV63 W5PKN2 K9J6A9 A0A2J8WBA4 B4LVB1 A0A3Q1M9F3 A0A250Y7S9 A0A3Q0D4E5 L8IGZ3 A0A3Q7NND3 Q3UL43 Q6ZQ45 A0A2R8VHH1 G1SCR7 Q99P88 A0A0G2K7T6 A0A3Q7NPA4 A0A2K5CJJ5 A0A024R071 O75694 F7CDY1 G3RZ49 A0A1S3AAS7 A0A2Y9QHF8 U3DWT8 K7AP62 A0A091CYW6 A0A2K6RRI9 U3FK31 Q80X48 A0A2K5X0U2 A0A1D5QL31 A0A2K5XFG1 G7P7D4 A0A2K5RME6 G7MPR2 A0A2U3XG59 M3YTX4 G5BTT4 A0A2U4AXJ5 U6CQH1 Q7ZWL0 I3MP94 A0A3Q3A962 A0A340WH03 A0A2Y9HJX2
Pubmed
26354079
22118469
19121390
20075255
28648823
24508170
+ More
30249741 21347285 20566863 20798317 25348373 23537049 18563158 23254933 18362917 19820115 17994087 28004739 17550304 22936249 17510324 12364791 21993624 9732281 10511559 10731132 12537572 10731138 17410542 17277377 21696798 22718353 26341556 20809919 19393038 28087693 28071753 22751099 10349636 11042159 11076861 11217851 12466851 16141073 14621295 19468303 21183079 19070573 23641069 15057822 15632090 11181995 10191094 12034489 9872452 15489334 17974005 14645504 15703211 18669648 19413330 20068231 21269460 21406692 22223895 22814378 23186163 24275569 28112733 19892987 22398555 25243066 16136131 25362486 17431167 22002653 25319552 21993625 27762356
30249741 21347285 20566863 20798317 25348373 23537049 18563158 23254933 18362917 19820115 17994087 28004739 17550304 22936249 17510324 12364791 21993624 9732281 10511559 10731132 12537572 10731138 17410542 17277377 21696798 22718353 26341556 20809919 19393038 28087693 28071753 22751099 10349636 11042159 11076861 11217851 12466851 16141073 14621295 19468303 21183079 19070573 23641069 15057822 15632090 11181995 10191094 12034489 9872452 15489334 17974005 14645504 15703211 18669648 19413330 20068231 21269460 21406692 22223895 22814378 23186163 24275569 28112733 19892987 22398555 25243066 16136131 25362486 17431167 22002653 25319552 21993625 27762356
EMBL
KQ460398
KPJ15054.1
KQ459606
KPI91439.1
AGBW02013719
OWR42917.1
+ More
NWSH01000130 PCG79234.1 ODYU01005260 SOQ45936.1 BABH01030920 BABH01030921 GBYB01003356 GBYB01013329 JAG73123.1 JAG83096.1 AAZX01000576 NNAY01000426 OXU28488.1 KK107997 QOIP01000013 EZA46992.1 RLU15247.1 ADTU01021605 KQ978273 KYM95793.1 KQ982691 KYQ52258.1 KQ976641 KYM78767.1 KQ980765 KYN13411.1 DS235366 EEB15317.1 KQ981744 KYN36228.1 NEVH01008206 PNF34664.1 GL442888 EFN62815.1 GFDL01014698 JAV20347.1 DS231817 EDS39688.1 GAKP01007794 JAC51158.1 UFQS01000266 UFQT01000266 SSX02233.1 SSX22610.1 SSX02235.1 SSX22612.1 GL450241 EFN81296.1 GANO01000575 JAB59296.1 KB631760 ERL85886.1 KQ414705 KOC63217.1 GG666529 EEN58615.1 KZ288191 PBC34501.1 KQ435687 KOX81376.1 GECZ01001684 JAS68085.1 KB202752 ESO88145.1 KQ971314 EEZ97868.2 CH902620 EDV32454.1 UFQT01002397 SSX33365.1 GEZM01030665 JAV85435.1 CH954177 EDV58941.1 CM000157 EDW88030.1 GEDC01026829 JAS10469.1 CM002910 KMY89676.1 CH477229 EAT47017.1 CH480818 EDW52541.1 AAAB01008964 EAA12437.4 AAPE02020438 AAPE02020439 AAPE02020440 APGK01009472 APGK01009473 APGK01009474 APGK01009475 APGK01009476 APGK01009477 APGK01009478 APGK01009479 KB737952 ENN82823.1 ABGA01167514 ABGA01167515 ABGA01167516 ABGA01167517 ABGA01167518 ABGA01167519 ABGA01167520 ABGA01167521 ABGA01167522 ABGA01167523 Y17111 AF051397 AF051398 AF051396 AE014134 AF160944 AAC05385.1 AAC05386.1 AAC06247.1 AAC06248.1 AAD46884.1 AAF53051.1 CAA76635.1 KQ759893 OAD62095.1 AMGL01035186 GABZ01000296 JAA53229.1 NDHI03003396 PNJ67043.1 CH940649 EDW63290.1 GFFV01000356 JAV39589.1 JH881428 ELR54397.1 AK145717 BAE26607.1 AK129217 BAC98027.1 AC107747 AAGW02030447 AAGW02030448 AAGW02030449 AAGW02030450 AF322375 AABR07008413 AABR07008414 CH474048 EDL75690.1 CH471119 EAW55958.1 AJ007558 AF165926 AB018334 BC039257 AL117585 CABD030105545 CABD030105546 CABD030105547 CABD030105548 CABD030105549 CABD030105550 CABD030105551 CABD030105552 GAMS01005320 JAB17816.1 AACZ04053631 AACZ04053632 GABC01007734 GABF01008724 GABD01004593 GABE01005623 NBAG03000430 JAA03604.1 JAA13421.1 JAA28507.1 JAA39116.1 PNI25611.1 KN123819 KFO23085.1 GAMT01005065 GAMR01003352 GAMQ01001821 GAMP01004097 JAB06796.1 JAB30580.1 JAB40030.1 JAB48658.1 BC050919 AAH50919.1 AQIA01058446 AQIA01058447 AQIA01058448 AQIA01058449 AQIA01058450 JSUE03035764 JSUE03035765 JSUE03035766 JSUE03035767 JSUE03035768 JSUE03035769 JSUE03035770 JSUE03035771 JSUE03035772 JSUE03035773 CM001281 EHH54205.1 JU321860 JU472862 JV044517 CM001256 AFE65616.1 AFH29666.1 AFI34588.1 EHH18476.1 AEYP01001369 AEYP01001370 AEYP01001371 AEYP01001372 AEYP01001373 AEYP01001374 AEYP01001375 AEYP01001376 AEYP01001377 AEYP01001378 JH171771 GEBF01000601 EHB12695.1 JAO03032.1 HAAF01000733 CCP72559.1 BC047162 CM004466 AAH47162.1 OCU02591.1 AGTP01040422 AGTP01040423
NWSH01000130 PCG79234.1 ODYU01005260 SOQ45936.1 BABH01030920 BABH01030921 GBYB01003356 GBYB01013329 JAG73123.1 JAG83096.1 AAZX01000576 NNAY01000426 OXU28488.1 KK107997 QOIP01000013 EZA46992.1 RLU15247.1 ADTU01021605 KQ978273 KYM95793.1 KQ982691 KYQ52258.1 KQ976641 KYM78767.1 KQ980765 KYN13411.1 DS235366 EEB15317.1 KQ981744 KYN36228.1 NEVH01008206 PNF34664.1 GL442888 EFN62815.1 GFDL01014698 JAV20347.1 DS231817 EDS39688.1 GAKP01007794 JAC51158.1 UFQS01000266 UFQT01000266 SSX02233.1 SSX22610.1 SSX02235.1 SSX22612.1 GL450241 EFN81296.1 GANO01000575 JAB59296.1 KB631760 ERL85886.1 KQ414705 KOC63217.1 GG666529 EEN58615.1 KZ288191 PBC34501.1 KQ435687 KOX81376.1 GECZ01001684 JAS68085.1 KB202752 ESO88145.1 KQ971314 EEZ97868.2 CH902620 EDV32454.1 UFQT01002397 SSX33365.1 GEZM01030665 JAV85435.1 CH954177 EDV58941.1 CM000157 EDW88030.1 GEDC01026829 JAS10469.1 CM002910 KMY89676.1 CH477229 EAT47017.1 CH480818 EDW52541.1 AAAB01008964 EAA12437.4 AAPE02020438 AAPE02020439 AAPE02020440 APGK01009472 APGK01009473 APGK01009474 APGK01009475 APGK01009476 APGK01009477 APGK01009478 APGK01009479 KB737952 ENN82823.1 ABGA01167514 ABGA01167515 ABGA01167516 ABGA01167517 ABGA01167518 ABGA01167519 ABGA01167520 ABGA01167521 ABGA01167522 ABGA01167523 Y17111 AF051397 AF051398 AF051396 AE014134 AF160944 AAC05385.1 AAC05386.1 AAC06247.1 AAC06248.1 AAD46884.1 AAF53051.1 CAA76635.1 KQ759893 OAD62095.1 AMGL01035186 GABZ01000296 JAA53229.1 NDHI03003396 PNJ67043.1 CH940649 EDW63290.1 GFFV01000356 JAV39589.1 JH881428 ELR54397.1 AK145717 BAE26607.1 AK129217 BAC98027.1 AC107747 AAGW02030447 AAGW02030448 AAGW02030449 AAGW02030450 AF322375 AABR07008413 AABR07008414 CH474048 EDL75690.1 CH471119 EAW55958.1 AJ007558 AF165926 AB018334 BC039257 AL117585 CABD030105545 CABD030105546 CABD030105547 CABD030105548 CABD030105549 CABD030105550 CABD030105551 CABD030105552 GAMS01005320 JAB17816.1 AACZ04053631 AACZ04053632 GABC01007734 GABF01008724 GABD01004593 GABE01005623 NBAG03000430 JAA03604.1 JAA13421.1 JAA28507.1 JAA39116.1 PNI25611.1 KN123819 KFO23085.1 GAMT01005065 GAMR01003352 GAMQ01001821 GAMP01004097 JAB06796.1 JAB30580.1 JAB40030.1 JAB48658.1 BC050919 AAH50919.1 AQIA01058446 AQIA01058447 AQIA01058448 AQIA01058449 AQIA01058450 JSUE03035764 JSUE03035765 JSUE03035766 JSUE03035767 JSUE03035768 JSUE03035769 JSUE03035770 JSUE03035771 JSUE03035772 JSUE03035773 CM001281 EHH54205.1 JU321860 JU472862 JV044517 CM001256 AFE65616.1 AFH29666.1 AFI34588.1 EHH18476.1 AEYP01001369 AEYP01001370 AEYP01001371 AEYP01001372 AEYP01001373 AEYP01001374 AEYP01001375 AEYP01001376 AEYP01001377 AEYP01001378 JH171771 GEBF01000601 EHB12695.1 JAO03032.1 HAAF01000733 CCP72559.1 BC047162 CM004466 AAH47162.1 OCU02591.1 AGTP01040422 AGTP01040423
Proteomes
UP000053240
UP000053268
UP000007151
UP000218220
UP000005204
UP000002358
+ More
UP000215335 UP000053097 UP000279307 UP000005205 UP000078542 UP000075809 UP000078540 UP000078492 UP000009046 UP000078541 UP000235965 UP000000311 UP000002320 UP000008237 UP000030742 UP000053825 UP000001554 UP000242457 UP000085678 UP000053105 UP000030746 UP000007266 UP000007801 UP000192223 UP000192221 UP000008711 UP000002282 UP000008820 UP000001292 UP000007062 UP000005203 UP000001074 UP000019118 UP000001595 UP000000803 UP000192224 UP000002356 UP000008792 UP000009136 UP000189706 UP000286641 UP000000589 UP000001811 UP000002494 UP000233020 UP000005640 UP000002281 UP000001519 UP000079721 UP000248483 UP000002277 UP000028990 UP000233200 UP000008225 UP000233100 UP000006718 UP000233140 UP000009130 UP000233040 UP000245341 UP000000715 UP000006813 UP000245320 UP000186698 UP000005215 UP000264800 UP000265300 UP000248481
UP000215335 UP000053097 UP000279307 UP000005205 UP000078542 UP000075809 UP000078540 UP000078492 UP000009046 UP000078541 UP000235965 UP000000311 UP000002320 UP000008237 UP000030742 UP000053825 UP000001554 UP000242457 UP000085678 UP000053105 UP000030746 UP000007266 UP000007801 UP000192223 UP000192221 UP000008711 UP000002282 UP000008820 UP000001292 UP000007062 UP000005203 UP000001074 UP000019118 UP000001595 UP000000803 UP000192224 UP000002356 UP000008792 UP000009136 UP000189706 UP000286641 UP000000589 UP000001811 UP000002494 UP000233020 UP000005640 UP000002281 UP000001519 UP000079721 UP000248483 UP000002277 UP000028990 UP000233200 UP000008225 UP000233100 UP000006718 UP000233140 UP000009130 UP000233040 UP000245341 UP000000715 UP000006813 UP000245320 UP000186698 UP000005215 UP000264800 UP000265300 UP000248481
Interpro
IPR014908
Nucleoporin_Nup133/Nup155_N
+ More
IPR017593 Allantoinase
IPR011059 Metal-dep_hydrolase_composite
IPR004870 Nucleoporin_Nup155
IPR006680 Amidohydro-rel
IPR007187 Nucleoporin_Nup133/Nup155_C
IPR032466 Metal_Hydrolase
IPR002195 Dihydroorotase_CS
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR011044 Quino_amine_DH_bsu
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR000719 Prot_kinase_dom
IPR011047 Quinoprotein_ADH-like_supfam
IPR018391 PQQ_beta_propeller_repeat
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR017593 Allantoinase
IPR011059 Metal-dep_hydrolase_composite
IPR004870 Nucleoporin_Nup155
IPR006680 Amidohydro-rel
IPR007187 Nucleoporin_Nup133/Nup155_C
IPR032466 Metal_Hydrolase
IPR002195 Dihydroorotase_CS
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR011044 Quino_amine_DH_bsu
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR000719 Prot_kinase_dom
IPR011047 Quinoprotein_ADH-like_supfam
IPR018391 PQQ_beta_propeller_repeat
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A194RC55
A0A194PFG3
A0A212EN81
A0A2A4K5N8
A0A2H1VZ07
H9JBB8
+ More
A0A0C9RA96 K7IWU3 A0A232FDD8 A0A026VTC4 A0A158NNH4 A0A151I9Q5 A0A151WWN5 A0A195B3A7 A0A195DKN5 E0VPL1 A0A195F702 A0A2J7R1G1 E2AUU0 A0A1Q3EYF6 B0W0E9 A0A034WBK6 A0A336LXS5 A0A336KFW9 E2BSQ0 W4VRP7 U4U5L9 A0A0L7QX73 C3YM21 A0A2A3ETX5 A0A1S3J6G6 A0A0M9AD09 A0A1B6H098 V3ZZP8 D6WBT9 B3MJZ9 A0A1W4WA12 A0A1W4W511 A0A336MUH0 A0A1Y1MLT3 B3N4Z6 B4P155 A0A1B6CAW0 A0A0J9R0K0 Q17JY3 B4HWZ6 Q7Q3R7 A0A088A0M9 G1PPP5 N6TXK7 H2PFD5 Q9V463 A0A1W4ZT04 A0A1W4ZS22 A0A310SV63 W5PKN2 K9J6A9 A0A2J8WBA4 B4LVB1 A0A3Q1M9F3 A0A250Y7S9 A0A3Q0D4E5 L8IGZ3 A0A3Q7NND3 Q3UL43 Q6ZQ45 A0A2R8VHH1 G1SCR7 Q99P88 A0A0G2K7T6 A0A3Q7NPA4 A0A2K5CJJ5 A0A024R071 O75694 F7CDY1 G3RZ49 A0A1S3AAS7 A0A2Y9QHF8 U3DWT8 K7AP62 A0A091CYW6 A0A2K6RRI9 U3FK31 Q80X48 A0A2K5X0U2 A0A1D5QL31 A0A2K5XFG1 G7P7D4 A0A2K5RME6 G7MPR2 A0A2U3XG59 M3YTX4 G5BTT4 A0A2U4AXJ5 U6CQH1 Q7ZWL0 I3MP94 A0A3Q3A962 A0A340WH03 A0A2Y9HJX2
A0A0C9RA96 K7IWU3 A0A232FDD8 A0A026VTC4 A0A158NNH4 A0A151I9Q5 A0A151WWN5 A0A195B3A7 A0A195DKN5 E0VPL1 A0A195F702 A0A2J7R1G1 E2AUU0 A0A1Q3EYF6 B0W0E9 A0A034WBK6 A0A336LXS5 A0A336KFW9 E2BSQ0 W4VRP7 U4U5L9 A0A0L7QX73 C3YM21 A0A2A3ETX5 A0A1S3J6G6 A0A0M9AD09 A0A1B6H098 V3ZZP8 D6WBT9 B3MJZ9 A0A1W4WA12 A0A1W4W511 A0A336MUH0 A0A1Y1MLT3 B3N4Z6 B4P155 A0A1B6CAW0 A0A0J9R0K0 Q17JY3 B4HWZ6 Q7Q3R7 A0A088A0M9 G1PPP5 N6TXK7 H2PFD5 Q9V463 A0A1W4ZT04 A0A1W4ZS22 A0A310SV63 W5PKN2 K9J6A9 A0A2J8WBA4 B4LVB1 A0A3Q1M9F3 A0A250Y7S9 A0A3Q0D4E5 L8IGZ3 A0A3Q7NND3 Q3UL43 Q6ZQ45 A0A2R8VHH1 G1SCR7 Q99P88 A0A0G2K7T6 A0A3Q7NPA4 A0A2K5CJJ5 A0A024R071 O75694 F7CDY1 G3RZ49 A0A1S3AAS7 A0A2Y9QHF8 U3DWT8 K7AP62 A0A091CYW6 A0A2K6RRI9 U3FK31 Q80X48 A0A2K5X0U2 A0A1D5QL31 A0A2K5XFG1 G7P7D4 A0A2K5RME6 G7MPR2 A0A2U3XG59 M3YTX4 G5BTT4 A0A2U4AXJ5 U6CQH1 Q7ZWL0 I3MP94 A0A3Q3A962 A0A340WH03 A0A2Y9HJX2
PDB
5IJO
E-value=0,
Score=1862
Ontologies
KEGG
GO
GO:0050897
GO:0017056
GO:0000256
GO:0008270
GO:0005643
GO:0004038
GO:0005524
GO:0004672
GO:0036228
GO:0006606
GO:0044611
GO:0000972
GO:0030715
GO:0034399
GO:0007283
GO:0030717
GO:0045477
GO:0035220
GO:0003682
GO:0005829
GO:0042307
GO:1905475
GO:0055050
GO:0007015
GO:0007303
GO:0001654
GO:1905938
GO:0006998
GO:0086014
GO:0031965
GO:0006406
GO:1905879
GO:0007295
GO:0015031
GO:0048477
GO:0001555
GO:0005634
GO:1900182
GO:0051028
GO:0005694
GO:0005737
GO:0007140
GO:0005635
GO:0007281
GO:0046930
GO:0015288
GO:0016925
GO:1900034
GO:0016032
GO:0043657
GO:0075733
GO:0016020
GO:0006110
GO:0060964
GO:0005215
GO:0006409
GO:0019083
GO:0005515
GO:0006508
GO:0004180
GO:0006207
GO:0046872
GO:0004088
GO:0006541
GO:0016812
GO:0016597
PANTHER
Topology
Subcellular location
Nucleus
Increased localization to chromatin before interphase (PubMed:22718353). Distributed along filamentous structures that extend radially on the nuclear side of the pore complex (PubMed:17410542). With evidence from 3 publications.
Nuclear pore complex Increased localization to chromatin before interphase (PubMed:22718353). Distributed along filamentous structures that extend radially on the nuclear side of the pore complex (PubMed:17410542). With evidence from 3 publications.
Chromosome Increased localization to chromatin before interphase (PubMed:22718353). Distributed along filamentous structures that extend radially on the nuclear side of the pore complex (PubMed:17410542). With evidence from 3 publications.
Nucleus membrane Increased localization to chromatin before interphase (PubMed:22718353). Distributed along filamentous structures that extend radially on the nuclear side of the pore complex (PubMed:17410542). With evidence from 3 publications.
Cytoplasm Increased localization to chromatin before interphase (PubMed:22718353). Distributed along filamentous structures that extend radially on the nuclear side of the pore complex (PubMed:17410542). With evidence from 3 publications.
Nuclear pore complex Increased localization to chromatin before interphase (PubMed:22718353). Distributed along filamentous structures that extend radially on the nuclear side of the pore complex (PubMed:17410542). With evidence from 3 publications.
Chromosome Increased localization to chromatin before interphase (PubMed:22718353). Distributed along filamentous structures that extend radially on the nuclear side of the pore complex (PubMed:17410542). With evidence from 3 publications.
Nucleus membrane Increased localization to chromatin before interphase (PubMed:22718353). Distributed along filamentous structures that extend radially on the nuclear side of the pore complex (PubMed:17410542). With evidence from 3 publications.
Cytoplasm Increased localization to chromatin before interphase (PubMed:22718353). Distributed along filamentous structures that extend radially on the nuclear side of the pore complex (PubMed:17410542). With evidence from 3 publications.
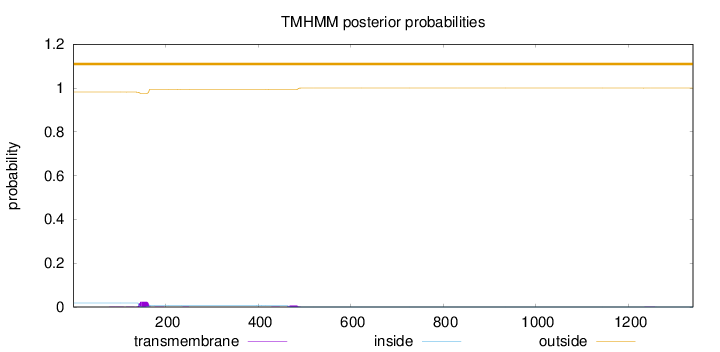
Length:
1339
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.677829999999996
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01803
outside
1 - 1339
Population Genetic Test Statistics
Pi
301.182819
Theta
190.532429
Tajima's D
1.258134
CLR
0.062509
CSRT
0.726463676816159
Interpretation
Uncertain
