Gene
KWMTBOMO05732
Pre Gene Modal
BGIBMGA006873
Annotation
PREDICTED:_neural_cell_adhesion_molecule_2-like_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.248 Nuclear Reliability : 1.057 PlasmaMembrane Reliability : 1.139
Sequence
CDS
ATGCATGCACGTAAATTAACTGTAACGTTGACCCAATTGTGTTGTGTGACAATGAACCGCATAGTGCATATCGTAGTTTTAGGTGTACTACTATTGACGTGCTGTATAGCCGACGAAGTTATTGAAACAGTAGAAGTCATTGTGGGACACAGCGCAGAGCTTCCTTGCAATGTCACAGCCAAGAACGAGAATGACAGACTAAGCATATTGGCCTGGTATCGTAATGAATCTGCGGTGGCTTTCTATAGTCGCGACCTCCGTGACGGTGCAGAAAACAGCAACCCATCTCCCGACAGTCGATACCGACTTGTTACTTCAGACTCTGAAGTCATGGATAAGCTGCAGATTGTAACAGTACGACCATCAGATGCGGGTCTCTATCACTGCCTCGCGGACTTCGAGTTCAGCCCTGCTCACAAGACTAACGTGCAGTTAGTGGTTATTGAGCCTCCACGTCACTTGTGGGTGATCCACGAGAACGGAACTCAGGTAGCTAAAGCAAGCGCAGGAGTCAACGTCAGTAGGAATGTGGGACCATACTACGTGGGAGATACAGTACATCTATTTTGTGTCGCCTTTGGTGGGAACCCTCAGGCGACACTATCCTGGTGGACAGAACAACGCCTCCTGAAGAACTCATCGACAACACTCTCAGAGCAACGAGTTCGCACTAACATAATGTTCGGTCCTCTCCGGCGTGAAGACCACGGCCGCGTACTCACCTGCTACGCAAAAAATAATGAAAGAACGAAGCCTCTTTCAATAGACATTATCATAAACATGCTCTTGCCTCCGGAACTTGTATCTATACGGGCTGAGGGGGCGTTTACTTCGGGCGGCGTTGAAGGCAGAGTGCGAACTGGAGAAGCCATAACATTGCAGTGCAGAGTAATTGGGGCACACCCCCCACCCCCACTGCTGTGGAAGCTTGACGATTCGAAGCTTGTAAATTTAGAGCAGAACATAACAATGGAGCCTTCGCAGCGCTTAATAGTTAGTGAGGTCCAATGGACAATAGACCGTCGGTACGACGAATCCCGCGTGACGTGCTGTGCCCTCAACTACCAAAGAGGAAATGACACCTTCGAATGCGCACAAGGAATACCTCTGATTGTTTTGTACCCTCCAGTTGTAGAGATTATTGTTGAAGGAGAATTAAATAATAATACTTTGGCTGTTGCAAAAGGCTCTAACGTAACGCTTGCTTGTAGCTATCTAGCTAATCCATCTGTGGACCAAATAATGTGGTTTCATGAGGATGCCGTACAAGGCAATGAAGGTGAGGTAACTGATAAACTACACCCGATTTTGGACCTGAAGAATATATCTGAGAGCGACGCCGGCAAATACATGTGTACCGCACATAACGAAGAGGGGTTCACAGACAGCGAACCGATTTTTATAGATGTCACGTATCCAGCTTACTGTGAAAATTCAATGATAATAGAATACGGTTTGGGCGATGAGGAGTCTATAAATATGACGTGCAATGTCAAAGCAAATCCAGAGCCAATCTCATACCGTTGGATTTCGGTCAACGCAGCCGCGAACATCACTACGCTGCGCAACCATCCCCAGGTTATGAAGGAAACAGATGAACCAATATTGGTTTATGAACGACCAAATAACACAGCGTACAGTACAGTATTTTGTTGGGGTCTCAACGGTGTGCCTGATAAGGAAGCCACGCAAACCCCTTGCACGTATTTAGTAACAGATGAAACGATCCCACGACCGCCTTCTAATTGTGAAGCTTTGATAACTGAAACAAGAGACATTATTGTTAACTGTGAGAAGGGTCACAGCGGGGGATTGCCACAGCGCTTCAAGTTTTTCGTGAAAGAGGAAGACACAGATAAACTCATAATATCCATTGTGAGTACAGAACCGGAATTTAAAATAGCGGAACCGAAGCAAGAAAGATACAAGTTTACTGTAATTGCAGTAAATGACAAAGGGGAAAGTGATAGTGTAGAAATTCTTAAAGAAGACATCATTAATAACATTCCAGACCCGGATCAGCCCCAGAGCGCTGTTGCGAACATAACGACGCTATCCCTGGCGCTGTGCGGCGGCGTGGCGCTAGTGGCGCTGGCGGCGTGCGGGCTCGTGCTGTGCGCGCACGACCGCGGCTCCGCGCTCGACCTGCCGCGGCCGCACTCGGACCCGCCGCTCTGCGCCTACAATACTGAAGAATCGAACTGCGAGACTTACCACGGCAGCGAAGACGGGAGCGAGTGCAACGTGAGACGCACGGAGTCGTTCCGGCGAGCCGTCTCGAGGTACCCGTCGAAGAATTTCGACGTGAGGAGAACGAGCTCGTTCCATTCGGCGCGCTACGCGAATGATACCGAGCCAGAACCTTCGAGCAAGTGCAACGACGTCATCCGCTACGGGAACAACTGCAGAGTGCACTCGCTACAGAACATCAGCAGGAAGAGGGACACGGATGCCTTGTGCGATCATTTAGTTCTACATCTGCCGCCGGAAACGAGCTACGGTGTTCCTAAGCCGATGAACACGTTTTACACGATACCGCGTAAAACGAGGCAAAAACAAAGCAGGGAACTGAGCGACGAGACTTCCGAAATAACTCAAACGTCCGACGGATTTTCGTTGCCGCCCCCGCCGGACGAGTTCGGGACCTACAGGGCCGCAACCAGGATCCGGGATATACCGAAAGCCACGCCTACATACACGACGGTCGTAAAGAAGAACTCCACCGGAAAGGAGCCAGTCAAATATCAACAGTACAACGATGCGATATCGTCAATGAACACTGTCGGAGTCGTGCCGACAATTAGCGCGACGCAAGCGAGCGTGTACACGTATCCTGACGACGACCACCGCGGGCACCAGGAAACGCCGAGCACTCTCGGAACCGCTAGCGTCATACCCACTTACAAAGCTTATTTTACCCCAAGAGAGTGCGGACTGTAA
Protein
MHARKLTVTLTQLCCVTMNRIVHIVVLGVLLLTCCIADEVIETVEVIVGHSAELPCNVTAKNENDRLSILAWYRNESAVAFYSRDLRDGAENSNPSPDSRYRLVTSDSEVMDKLQIVTVRPSDAGLYHCLADFEFSPAHKTNVQLVVIEPPRHLWVIHENGTQVAKASAGVNVSRNVGPYYVGDTVHLFCVAFGGNPQATLSWWTEQRLLKNSSTTLSEQRVRTNIMFGPLRREDHGRVLTCYAKNNERTKPLSIDIIINMLLPPELVSIRAEGAFTSGGVEGRVRTGEAITLQCRVIGAHPPPPLLWKLDDSKLVNLEQNITMEPSQRLIVSEVQWTIDRRYDESRVTCCALNYQRGNDTFECAQGIPLIVLYPPVVEIIVEGELNNNTLAVAKGSNVTLACSYLANPSVDQIMWFHEDAVQGNEGEVTDKLHPILDLKNISESDAGKYMCTAHNEEGFTDSEPIFIDVTYPAYCENSMIIEYGLGDEESINMTCNVKANPEPISYRWISVNAAANITTLRNHPQVMKETDEPILVYERPNNTAYSTVFCWGLNGVPDKEATQTPCTYLVTDETIPRPPSNCEALITETRDIIVNCEKGHSGGLPQRFKFFVKEEDTDKLIISIVSTEPEFKIAEPKQERYKFTVIAVNDKGESDSVEILKEDIINNIPDPDQPQSAVANITTLSLALCGGVALVALAACGLVLCAHDRGSALDLPRPHSDPPLCAYNTEESNCETYHGSEDGSECNVRRTESFRRAVSRYPSKNFDVRRTSSFHSARYANDTEPEPSSKCNDVIRYGNNCRVHSLQNISRKRDTDALCDHLVLHLPPETSYGVPKPMNTFYTIPRKTRQKQSRELSDETSEITQTSDGFSLPPPPDEFGTYRAATRIRDIPKATPTYTTVVKKNSTGKEPVKYQQYNDAISSMNTVGVVPTISATQASVYTYPDDDHRGHQETPSTLGTASVIPTYKAYFTPRECGL
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
4OFY
E-value=1.20569e-09,
Score=155
Ontologies
GO
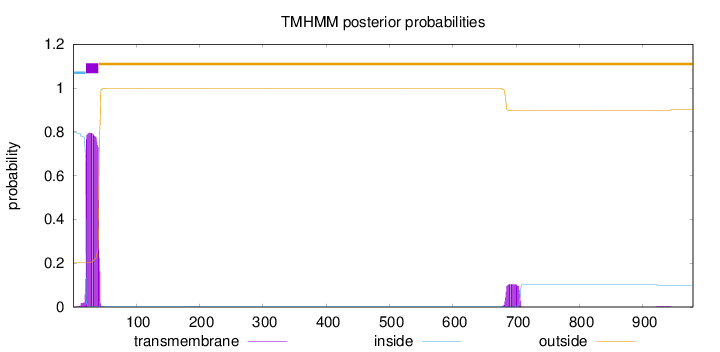
Topology
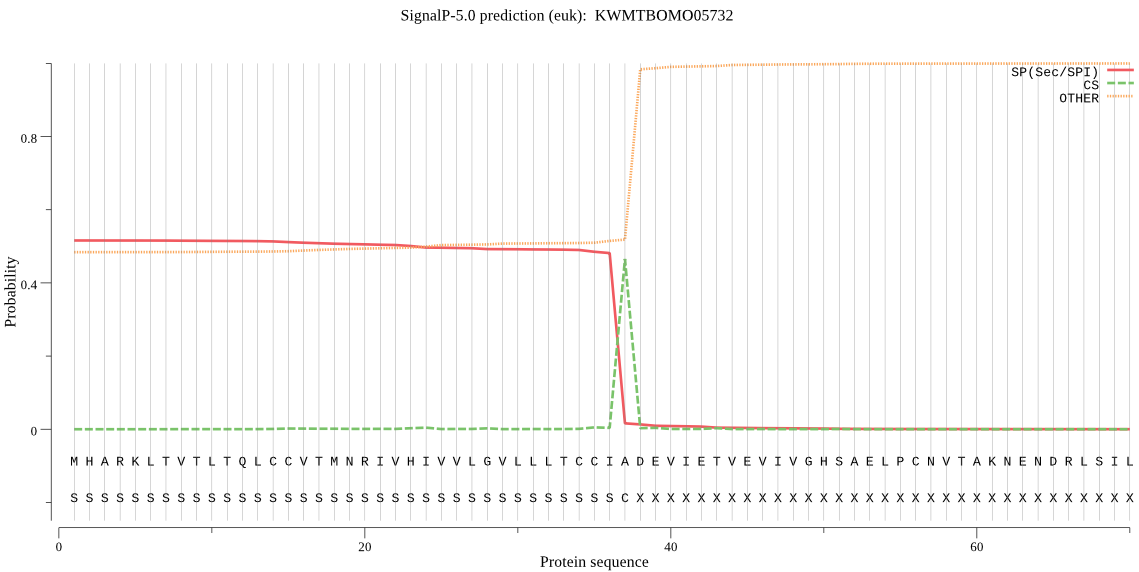
SignalP
Position: 1 - 37,
Likelihood: 0.516576
Length:
979
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.2645999999999
Exp number, first 60 AAs:
16.82541
Total prob of N-in:
0.79582
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 40
outside
41 - 979
Population Genetic Test Statistics
Pi
218.100753
Theta
176.043537
Tajima's D
0.509181
CLR
0.599788
CSRT
0.515774211289436
Interpretation
Uncertain
