Gene
KWMTBOMO05730
Pre Gene Modal
BGIBMGA006871
Annotation
Carboxypeptidase_A2_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.375 Nuclear Reliability : 1.249
Sequence
CDS
ATGGCGCGTCGCCTGGCGCTGCTCGCGCTGCTGGCGTGCGTGGTGGCGCTTCGCCGCCCGCCTCCTACTTACGACGACATGTACTACTCCGACTCCGGGGACTCCTATTCCCTGTGGGATGATTATGACGACTCCTACGAAACAACCCCTCCGCCGCGATTACGTCCTACGACGACTAGTGCTAAAGTGATACTAAGGAATAAAGTTCCAAAAGTGAACGTGACGGTTAAAACGTCAAAAGGTGATGATAAAAAAATAATAAAAACCACAAACATTAATATTGAGAGTCATACAAACTTTATAGTGACAAAGGCTCCTGTTGTGAGTCAAGGGACGGTCACAAGGCGCCCTGTGATCCAAATCAGTGAAGACTATGTTATAGTTGAATCTCCAGAGCTACATGCCGATCAGTTTTCAAAATCTAGCACGAAAAAGCCTGTGACGTCACGTCGGCCATCCTCCAGCAAGCGACCTACGACAAAGCGGCCCCCAACGACTCGTCGATCGCGACCACCGCTTAAAACTACCAAAGCTCCAAAAGTAAAAAAACCTACATGTCCCCCGAAAAGTACTGGGTGGCTGTCATCATTTTTTCAAGATGTCAAAGAAAAAAAAACTCCCAAGAAACCCGCAAAATCAGTTCCAGGTATATTTTCAAATATATACAATTGGTTTGTGAACAAAAAAACTTCATGGTTCGGCGGAGACGGCGGATGGTTCCAAGGTTCGGTAAAAGCCAAAAAACCAAATTATATTTTTCCAACGACAACCACACCAGAGCCGCTCACGCCTGCGCCCAGCTGGTTCCATACCCTTGTTGAATCGGAGGAAAACGATGGTGCCTACGCCAGCGAACTCAGTACAAGTAAGAAACCAACAAAGAAATCTTTGAAGAAGACATACAAAGGTTATCAGTTGTTGCGAACATACCCAGACGTACAATGGAAGGTTCAAGAAATGTTAGATTTACAAGAAGAAGCTCAAGGTTCTGGTATAATGTGGTGGACTCCACCCTCTCTGAATGGCACTACGGATTTAGTCGTACCCTCAGACTTACTAGAAGACGTCAAAGAACAGTTAAAATCATCGCAGATAGATTACGATGTAGTTATTTGGGACTTGCAGAAAGCCATCGCCTACGAAAATCCACGTTTATCAAAAAAACAAAGAATAGAACTTGAACAAGTTCGAGGACATCCGATGACCTGGAGACGTTATCATCGTTACGCCGACATATTACGTTATTTGGAGTATTTACAACATTCTCATTCCGATGTAATAGAACTGATTCCTCTCGGTCGCTCTTCCGAAGGTCTTCCTTTAGTGGCTGTTAAGATCTCGGTATCGAATGACAATCAGATGAAGGAATCTGGAAATAAAACAAATAAAAAGAAATGGAAGCTTCGGTCCCATATGAAACCAGCTGTGTGGATCGATGGAGGAACTCACGCTCGGGAGTGGATAGCACCGGCTGTCGCAACATGGATAATCCACACTCTTGTTGAAGGAGAAAAAGGCTTAGGAGCAGACTTAGAAGTACTAAAGCAAGCTGACTTCTATATAATGCCAGTGTTAAACCCAGACGGGTATGAACACTCGCACACGCATGACCGACTATGGAGCAAAACGCGTTCACGCTCCCCCGATCATTCTGACGACTACTTCGTTGGCTGGTTTCCATGGAACTGGGGTCAAGTTGAATGTGTGGGTGTGGATGCAGACCGCAACTGGGACTACCATTGGGGCGAGAAAGACTCCTCGACGGATCCTTGCTCAGATAACTACGCTGGGCCCCATCCGTTCAGTGAACCCGAGACGCGGGCTGTTTCAGAATTTCTTGCAGAGCATCGAGGACAAATTAAAGTTTTTATATCTCTGCATGCCTATACCCAAGCTTGGCTAGTACCAAGTTCCCAAGCCCACGCGCCCTTCGCCGATGATGGAATACTCATGGAAATGGGAAAATTGGCAACAACGGCGCTTTCCGACTTATTCGGTACCAAGTATCAGCTGGGCACTGTGGCAGATTTGCGGCAGCCTGCGTCTGGGATGTCTCATGATTGGGCCAAGGCTCGGGCTGGCATCAAGTTTTCCTATCACGTTGACTTGCGAGATTCGTTTGGACCTTACGGCTTTCTACTACCTGGATCGCAAATTGTATCAACGGCTCGCGAAACTTGGCAAGCAGTTAAAGCAATTGTGGACAATTTGACACCTTAA
Protein
MARRLALLALLACVVALRRPPPTYDDMYYSDSGDSYSLWDDYDDSYETTPPPRLRPTTTSAKVILRNKVPKVNVTVKTSKGDDKKIIKTTNINIESHTNFIVTKAPVVSQGTVTRRPVIQISEDYVIVESPELHADQFSKSSTKKPVTSRRPSSSKRPTTKRPPTTRRSRPPLKTTKAPKVKKPTCPPKSTGWLSSFFQDVKEKKTPKKPAKSVPGIFSNIYNWFVNKKTSWFGGDGGWFQGSVKAKKPNYIFPTTTTPEPLTPAPSWFHTLVESEENDGAYASELSTSKKPTKKSLKKTYKGYQLLRTYPDVQWKVQEMLDLQEEAQGSGIMWWTPPSLNGTTDLVVPSDLLEDVKEQLKSSQIDYDVVIWDLQKAIAYENPRLSKKQRIELEQVRGHPMTWRRYHRYADILRYLEYLQHSHSDVIELIPLGRSSEGLPLVAVKISVSNDNQMKESGNKTNKKKWKLRSHMKPAVWIDGGTHAREWIAPAVATWIIHTLVEGEKGLGADLEVLKQADFYIMPVLNPDGYEHSHTHDRLWSKTRSRSPDHSDDYFVGWFPWNWGQVECVGVDADRNWDYHWGEKDSSTDPCSDNYAGPHPFSEPETRAVSEFLAEHRGQIKVFISLHAYTQAWLVPSSQAHAPFADDGILMEMGKLATTALSDLFGTKYQLGTVADLRQPASGMSHDWAKARAGIKFSYHVDLRDSFGPYGFLLPGSQIVSTARETWQAVKAIVDNLTP
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
2BOA
E-value=2.09472e-61,
Score=600
Ontologies
GO
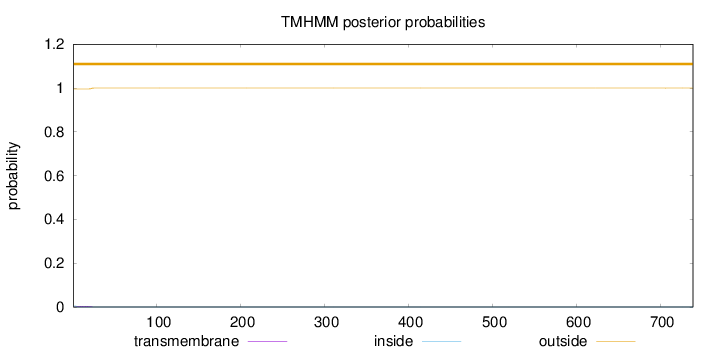
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.963644
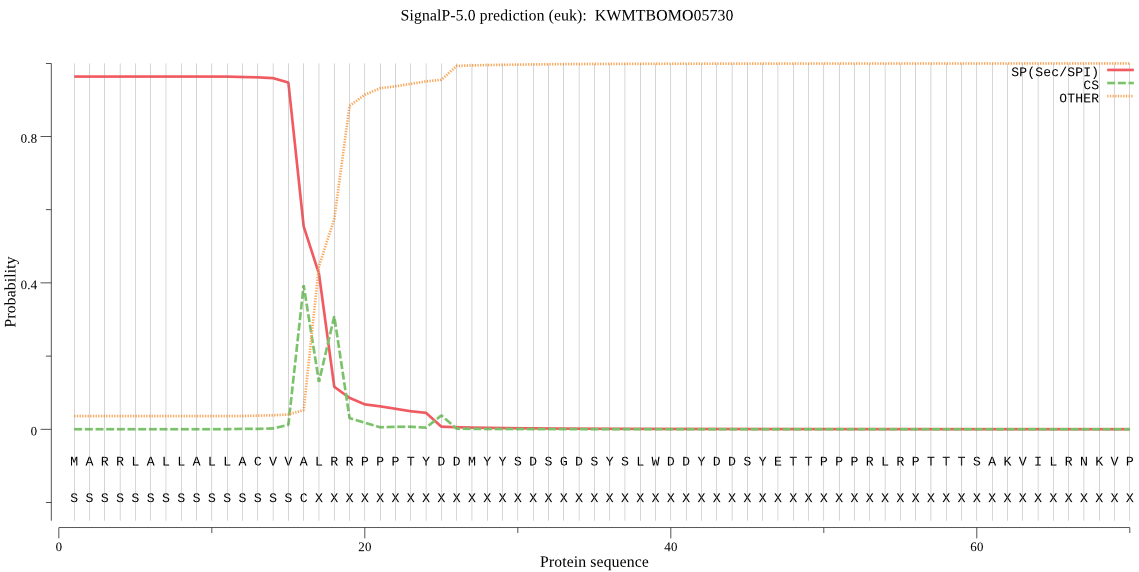
Length:
739
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0833099999999998
Exp number, first 60 AAs:
0.08092
Total prob of N-in:
0.00468
outside
1 - 739
Population Genetic Test Statistics
Pi
202.751558
Theta
193.605809
Tajima's D
0.097442
CLR
0.953264
CSRT
0.395680215989201
Interpretation
Uncertain
