Gene
KWMTBOMO05728 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006816
Annotation
PREDICTED:_CAD_protein_isoform_X1_[Amyelois_transitella]
Full name
Beta-galactosidase
+ More
CAD protein
CAD protein
Alternative Name
Protein rudimentary
Location in the cell
Cytoplasmic Reliability : 2.731
Sequence
CDS
ATGGAAAATGATGATTTGAGCTGCGACTTAGGCCCGCTGTGTTGCCTTGTGCTGGAGGATGGAACTATACTTCATGGTAGGAGCTTTGGTGCTCAAGTGCCCGTTGAAGGCGAAGTTGTTTTTCAAACGGGCATGGTAGGATATCCGGAATCCCTGACAGATCCGTCGTACAACGCACAACTACTGGTGTTAACTTATCCTCTCATTGGAAACTATGGAGTTCCGGATGAGAATGACCGAGACGAACACGGTATTAGCAGGTGGTTTGAATCCGACCGTATTTGGGCAGCTGGTTTGATAGTGGGGGAAGTCAGCAAACGTGCTTATCATTGGCGTGCCAAATGTTCCATAGGAGCATGGCTAGCCCAGCATCGGGTCCCTGGTATATGTGACATTGATACACGCGCTCTAACTTGCCGTCTCAGAGAAGGTATAACTCTCGGGAAAATCATACAAGGCTTACCGCCGTCTACTCCACTTCCACCGTTCACAGATCCGAACTTACGAAACTTAGTTGCCGAAGTGTCAATTAAGCAACCACGAGTTTTTAATAAAAATGGAGAGATAACAATTTTGGCAATCGACTGCGGTTCAAAATACAATCAAATTAGATGTTTTATCAAAAGGAACGCGAAAGTCGTTCTAGTACCGTGGGACTACAAGCCCAACCCAAATGAATACGATGGCTTGTTCATAAGCAACGGGCCAGGCGATCCGGAAATTTGCAAAAAAGTTGTAGAAAACTTACAATTTGTTATGAAAAATCAAAATATTGTAAAACCTATCTTTGGTATTTGTCTAGGACATCAACTATTATCTACCGCAGCGGGGTGCAAAACTTACAAGACCAAATATGGCAACCGTGGTCACAACTTGCCGTGTACTCACAGTTCCACTGGCAGATGTTTCATGACTTCGCAGAACCATGGCTACGCCGTCGATACCGATTCTTTGCCTGAAGGATGGAAAGTTTTGTTCACCAATGAAAATGATAAGACTAACGAAGGAATCATTCACAAGTCTGCGCCTTATTTCAGCGTTCAATTTCATCCTGAACACACTGCGGGCCCTACGGATTTAGAGTTTTTGTTTGACATTTTTGTTGATGCAGTTAAAAGTTATAAAAATAACGTAAATTGTGTTATAGATGAAATGATAAATGAAAAATTGAAATTTATTCCAACAATTCACGATAGACCGAAAAAAGTATTAATCCTAGGCTCCGGAGGCTTATCAATAGGACAAGCCGGAGAGTTCGATTATTCAGGTTCGCAAGGCGTGAAGGCTATGCAAGAAGAAAAAATTCTAACAGTTCTTATAAATCCTAACATTGCCACTGTACAAACTTCTAAAGGTTTGGCGGATAAAGTTTACTTTTTACCTATAACTCCTGAATATGTAGAACAAGTCATAAAAGCAGAAAGACCAACGGGGATTTTGCTCACTTTTGGCGGACAAACTGCACTAAATTGCGGAGTGGAACTACAAAAATCTAAAATATTTGAAAAATATAATGTCAGCGTTTTAGGCACACCCATTCAGTCGATAGTTGATACTGAAGATAGGAAGATATTTGCAGAGAAAATCAATGCGATAGGAGAAAAAGTGGCACCAAGTGCAGCAGTAACATCAGTCGAAGAAGCACTCGAAGCAGCTCTGCAAATCGGGTATCCGGTGATGGCTCGCTCAGCCTTTTCCTTGGGAGGATTAGGCTCTGGTTTTGCGAACAATGAAGATGAACTCAAAACATTAGCTCACCAAGCTCTTTCACATTCCGATCAACTCGTTATTGACAAATCTTTGAAGGGGTGGAAAGAAGTTGAATACGAAGTCGTAAGAGACGCTTATGATAACTGCATAACTGTTTGTAATATGGAAAACGTTGACCCATTAGGTATACACACTGGAGAATCTATAGTAGTAGCGCCAAGTCAAACTTTGTCTAATAGAGAGTATTATATGCTTCGAAATACGGCTATAAAAGTTATACGACATTTTGGTATAGTCGGCGAATGTAATATACAATACGCTTTAAACCCGAATTCTGAAGAATTTTACATTATTGAAGTAAATGCAAGACTTTCGAGGAGCTCAGCATTAGCCAGTAAAGCAACAGGTTATCCATTAGCATATGTGGCTGCAAAATTAGCTCTCGGAATATCCTTGCCTGTTATCAAAAATTCGGTTACTGGAGTAACCACAGCCTGTTTTGAACCAAGTTTGGACTACTGTGTTGTTAAAATCCCAAGATGGGACCTCGCCAAATTTAACAAAGTGAGTACGAAAATTGGAAGCTCTATGAAAAGTGTTGGGGAAGTCATGTCTATCGGCAGGACTTTCGAAGAAGCATTCCAAAAAGCTTTGCGGATGGTTGACGAGAATGTAAATGGTTTTGATCCGAACATCAAAAAAGTAAACGAAAATGAGTTAAAGGAACCAACAGATAAACGTATGTTTGTGTTGGCTGCAGCCTTAAAGAACGGATTTAGTGTTGAAAAGTTGTATGAGTTGACTAAAATAGACAAATGGTTCTTAGAAAAGTTTAAGAACATTATTGACTATTACAAAACTCTTGAAAAATTAGATTCTGGAACAATTACATCTGACATTTTAAAGCAAGCTAAAAAAATGGGATTCTCTGATAAACAAATTGCTTCTGCAATAAAGAGTACTGAAGTAGCGATCCGTAAATTAAGAGAAGAATTCAAAATAACACCATTTGTTAAACAAATAGATACAGTTGCTGCGGAGTGGCCAGCTACTACCAACTATTTGTATTTAACCTACAATGGAAGCACACATGACATTGAATTTCCGGGAGAATTCGTTATGGTTTTAGGTTCAGGGGTGTATAGAATAGGAAGTTCGGTAGAGTTCGATTGGTGTGCCGTAGGATGTCTCAGGGAATTAAGAAATCAAGGTAAAAAAACAATAATGATAAATTACAATCCTGAAACAGTTAGTACTGATTATGACATGAGCGATCGTCTCTATTTTGAGGAAATATCATTTGAAGTTGTAATGGATATTTATGATATTGAAAGACCTAGTGGTGTAATATTGTCTATGGGGGGACAGCTACCAAATAATATTGCTATGGACCTTCATAGGCAACAAGCAAAAATATTAGGTACATCGCCGGACATGATTGATGGCGCTGAAAACAGGTTTAAATTTTCGAGAATGTTAGACCGGAAAGGTATTATGCAACCAAGGTGGAAAGAACTTACGGATTTAGAATCGGCTATTGGATTTTGTGAGGAAGTGGGATACCCATGCTTAGTGCGTCCATCGTACGTTTTAAGCGGTGCTGCAATGAATGTGGCTAATTCTCATCAAGAGTTAGAAACATATTTGAAATCTGCAAGTCAGTTAAGTAAAGAACATCCAGTAGTAATTTCTAAATTCATTATGGATGCAAAAGAAATAGACGTAGACGTTGTAGCAGCTGATGGAATAATTTATTGCATGGCTGTTTCCGAACATGTTGAAAACGCCGGTGTGCACTCTGGAGACGCAACCCTAGTAACCCCGCCACAGGATATTAACAACGAAACTTTAGAAAAGATAAAAGAAATATCTGGAATCATAGCTGAAACTTTAGACGTAACTGGTCCATTTAATATGCAACTGATAGCAAAAGATAATGACCTAAAAGTTATCGAATGCAACGTTAGGGTATCGAGATCTTTTCCATTTGTGTCTAAAACATTAGATCATGATTTTGTGGCAATGGCCACTAAAATTATTCTGGGACTACCAGTTGAACCGGCTAATGTGATGGCGGGATGTGGAAAGGTTGGAGTAAAAGTTCCACAATTTTCATTTTCAAGACTGGCCGGTGCAGACGTTACATTAGGTGTGGAAATGGCATCTACTGGAGAAGTTGCATGTTTTGGAGAAAATCGATACGAGGCATACTTAAAATCGTTAATGAGTACAGGCTTCAGGATTCCAAAAAAAGCTATTTTATTGTCTATAGGAACATTCAAGCATAAAATGGAATTATTGCCGAGCGTTCGATCGTTACAAAAAATGGGATATAAACTTTACGCTAGTATGGGTACTGGTGATTTTTATACAGAGCATGGAATCGAGATAGAGAGTGTCCAGTGGACATTTGAGCACATAGGAGATCCAGAAGATGATAGGTTTGACGGCGAACTAATGCACTTAGCCGATTTTATGGCTCGCCGACAATTAGATTTGGTGATCAATTTACCGATGCGAGGTGGTGCCCGTCGTGTTTCTTCATTTACTACGCACGGATACCGGACCCGGCGGCTCGCTGTTGATTATGCAGTACCTCTAGTGACCGATGTCAAATGCGCCAAGCTCTTAGTGCAGGCAATGTCACAATGTGGCGGTGCTCCTACAATGAAAACTCATACTGATTGTATGACGTCACGGAATATTGTCAAATTACCCGGCTTCATTGATGTACATGTTCACGTCCGTGAACCCGGTGCAACATATAAGGAAGATTTTGAGACCTGTACCGCAGCCGCATTGGCTGGTGGTGTAACGATGATTTGTGCTATGCCCAACACTAACCCCTCAATCGTTGATCGTACAGCATTTGATTACGCTTCAACATTAGCAAGAGTTAGTGCCCGATGCGATTATGCTTTGTACGTCGGTGCTTCCTCGACAAATTATGACACAATATGTGAACTGGCTCCGCAAGCTGCAGCTCTAAAGATGTACTTAAATGAAACTTTTACAACTCTTCTGTTAGAGGATATGACAATTTGGCAGAAACATTTCCAAAGTTGGCCCAAAAAAATGCCTGTTTGTACTCATGCAGAGCGTGAGAAAACCGCCGCTGTCATTTTAATGGCATCTCTCCTTGATCGGCCTGTACACATTTGTCACGTAGCACGAAAAGAAGAAATATTAATTATCAAAGCCGCAAAAGAAAGAGGTGTTAAAGTTACTTGTGAAGTTTGCCCACATCACTTATTTTTGAATAGTAATGACATTGACAAAATTGGAAAAGGGCGAGCAAAAGTATGTCCAGTATTATGTAGTCCTGAAGACCAAGCAGAACTTTGGCGAAATATAAGTATCATTGATGTTTTTGCCACAGACCATGCACCTCATACTGTGGAAGAAAAGAATTCTGAGAATCCACCACCTGGCTACCCTGGCTTAGAAACAATATTGCCTTTGCTACTCAACGCTGTACATCAAGGTCGTTTGACTATTGAAGATCTTATTAATAAATTTCATCGTAATCCTCGTAAAATTTTTAACTTACCAGAACAGCCTAACACATATGTTGAAGTTGATTTGGATTATGAATGGGTAATTCCAACGTCAATGGAATTTTCTAAATCGAAATGGACTCCTTTCGCGGGCATGAAGGTATGTGGTGCTATTCATCGTGTCACATTAAGAGGTGAAATAGCATATGTCGAAGGTCAAATTTTGGTTCCTCCAGGATTCGGTCAAAATGTTCGCGAATGGCCAATGCCTAAAAAACAAACTTTCCAGACGTACTCGATTGAAAAAGTTGAAAAAGAAATAAGTCGTCCTAATTCCGCATTGGATATAATTACACCTGGGGACCATCTAAGACAAAGCGATATTGACCTAGATCAATTGGAGTCTTCAAGATCAGAAGCTGTTGGAAGACTAAACGTGCATTTTAATGAAGTGGCCGGTATGCGAAACGTATCCCCTGTTCCACCACAAACAGTTACTAGACAAAGATGTGATAGCTCATCGAATTACAATCCACCACAGCCTTCAATTACACAACGACAACGCAGTGATTTGTTCGGCAAAAACATCCTGACTGTAGATACATTTAATAAGGAAACTCTCAACGACATTTTCAACCTAGCACAATTCATGAAGACAAGTGTAAATAAGGGACGTTATTTAGATGACATCTTACGTGGGAAAGTTATGGCCTCAATATTTTATGAAGTCAGTACACGTACGAGCTGCAGTTTTGCAGCGGCCATGCATAGACTAGGCGGATCTGTGGTACATACCGATGCAACAAGCTCTTCTGCAAAAAAAGGCGAATCTCTTGAAGATAGCGTCGCTATTATGGCTAGCTATTCAGATGTCGTCGTGTTGAGACATCCTGAACCCGGCGCAGTTTCTCGCGCATCGAAACTTAGTCGCAAGCCCGTAATAAATGCGGGAGACGGTGTTGGTGAACACCCCACGCAGGCATTGTTAGACGTTTTTACGATACGAGAAGAAATCGGTACAGTAAATGGCCTCACGATCACAATGGTTGGCGATCTTAAAAATGGACGCACAGTTCATTCCCTTGCTAGACTTTTAACTCTATATCAAGTCCAACTTCAATACGTAAGTCCGCCGGGACTAGGTATGCCGAAGCATATCACGGATTATGTTGCTTCTAAAGGTATACCTCAGAGAGTATACGAGCGCTTAGAGGACGTACTAGCTGATACTCACGTTTTGTACATGACTAGAATACAACGAGAAAGATTCAAAAGCCAAGAAGATTATGAAAAGACTCGGGGACTACTGGTTGTGACCCCACAGTTGATGACCAGGGCTCGTCGTCGTATGATAGTGATGCACCCTCTACCACGTGTCGATGAAATTTCATCAGAATTTGACTCAGATCCTCGCGCAGCATACTTTAGACAGGCCGAATACGGAATGTATGTTCGTATGGCTCTACTCGCAATGGTTGCTGGTGTAAATCCACTTGTGTGA
Protein
MENDDLSCDLGPLCCLVLEDGTILHGRSFGAQVPVEGEVVFQTGMVGYPESLTDPSYNAQLLVLTYPLIGNYGVPDENDRDEHGISRWFESDRIWAAGLIVGEVSKRAYHWRAKCSIGAWLAQHRVPGICDIDTRALTCRLREGITLGKIIQGLPPSTPLPPFTDPNLRNLVAEVSIKQPRVFNKNGEITILAIDCGSKYNQIRCFIKRNAKVVLVPWDYKPNPNEYDGLFISNGPGDPEICKKVVENLQFVMKNQNIVKPIFGICLGHQLLSTAAGCKTYKTKYGNRGHNLPCTHSSTGRCFMTSQNHGYAVDTDSLPEGWKVLFTNENDKTNEGIIHKSAPYFSVQFHPEHTAGPTDLEFLFDIFVDAVKSYKNNVNCVIDEMINEKLKFIPTIHDRPKKVLILGSGGLSIGQAGEFDYSGSQGVKAMQEEKILTVLINPNIATVQTSKGLADKVYFLPITPEYVEQVIKAERPTGILLTFGGQTALNCGVELQKSKIFEKYNVSVLGTPIQSIVDTEDRKIFAEKINAIGEKVAPSAAVTSVEEALEAALQIGYPVMARSAFSLGGLGSGFANNEDELKTLAHQALSHSDQLVIDKSLKGWKEVEYEVVRDAYDNCITVCNMENVDPLGIHTGESIVVAPSQTLSNREYYMLRNTAIKVIRHFGIVGECNIQYALNPNSEEFYIIEVNARLSRSSALASKATGYPLAYVAAKLALGISLPVIKNSVTGVTTACFEPSLDYCVVKIPRWDLAKFNKVSTKIGSSMKSVGEVMSIGRTFEEAFQKALRMVDENVNGFDPNIKKVNENELKEPTDKRMFVLAAALKNGFSVEKLYELTKIDKWFLEKFKNIIDYYKTLEKLDSGTITSDILKQAKKMGFSDKQIASAIKSTEVAIRKLREEFKITPFVKQIDTVAAEWPATTNYLYLTYNGSTHDIEFPGEFVMVLGSGVYRIGSSVEFDWCAVGCLRELRNQGKKTIMINYNPETVSTDYDMSDRLYFEEISFEVVMDIYDIERPSGVILSMGGQLPNNIAMDLHRQQAKILGTSPDMIDGAENRFKFSRMLDRKGIMQPRWKELTDLESAIGFCEEVGYPCLVRPSYVLSGAAMNVANSHQELETYLKSASQLSKEHPVVISKFIMDAKEIDVDVVAADGIIYCMAVSEHVENAGVHSGDATLVTPPQDINNETLEKIKEISGIIAETLDVTGPFNMQLIAKDNDLKVIECNVRVSRSFPFVSKTLDHDFVAMATKIILGLPVEPANVMAGCGKVGVKVPQFSFSRLAGADVTLGVEMASTGEVACFGENRYEAYLKSLMSTGFRIPKKAILLSIGTFKHKMELLPSVRSLQKMGYKLYASMGTGDFYTEHGIEIESVQWTFEHIGDPEDDRFDGELMHLADFMARRQLDLVINLPMRGGARRVSSFTTHGYRTRRLAVDYAVPLVTDVKCAKLLVQAMSQCGGAPTMKTHTDCMTSRNIVKLPGFIDVHVHVREPGATYKEDFETCTAAALAGGVTMICAMPNTNPSIVDRTAFDYASTLARVSARCDYALYVGASSTNYDTICELAPQAAALKMYLNETFTTLLLEDMTIWQKHFQSWPKKMPVCTHAEREKTAAVILMASLLDRPVHICHVARKEEILIIKAAKERGVKVTCEVCPHHLFLNSNDIDKIGKGRAKVCPVLCSPEDQAELWRNISIIDVFATDHAPHTVEEKNSENPPPGYPGLETILPLLLNAVHQGRLTIEDLINKFHRNPRKIFNLPEQPNTYVEVDLDYEWVIPTSMEFSKSKWTPFAGMKVCGAIHRVTLRGEIAYVEGQILVPPGFGQNVREWPMPKKQTFQTYSIEKVEKEISRPNSALDIITPGDHLRQSDIDLDQLESSRSEAVGRLNVHFNEVAGMRNVSPVPPQTVTRQRCDSSSNYNPPQPSITQRQRSDLFGKNILTVDTFNKETLNDIFNLAQFMKTSVNKGRYLDDILRGKVMASIFYEVSTRTSCSFAAAMHRLGGSVVHTDATSSSAKKGESLEDSVAIMASYSDVVVLRHPEPGAVSRASKLSRKPVINAGDGVGEHPTQALLDVFTIREEIGTVNGLTITMVGDLKNGRTVHSLARLLTLYQVQLQYVSPPGLGMPKHITDYVASKGIPQRVYERLEDVLADTHVLYMTRIQRERFKSQEDYEKTRGLLVVTPQLMTRARRRMIVMHPLPRVDEISSEFDSDPRAAYFRQAEYGMYVRMALLAMVAGVNPLV
Summary
Catalytic Activity
Hydrolysis of terminal non-reducing beta-D-galactose residues in beta-D-galactosides.
2 ATP + H2O + hydrogencarbonate + L-glutamine = 2 ADP + carbamoyl phosphate + 2 H(+) + L-glutamate + phosphate
carbamoyl phosphate + L-aspartate = H(+) + N-carbamoyl-L-aspartate + phosphate
(S)-dihydroorotate + H2O = H(+) + N-carbamoyl-L-aspartate
2 ATP + H2O + hydrogencarbonate + L-glutamine = 2 ADP + carbamoyl phosphate + 2 H(+) + L-glutamate + phosphate
carbamoyl phosphate + L-aspartate = H(+) + N-carbamoyl-L-aspartate + phosphate
(S)-dihydroorotate + H2O = H(+) + N-carbamoyl-L-aspartate
Cofactor
Zn(2+)
Miscellaneous
GATase (glutamine amidotransferase) and CPSase (carbamoyl phosphate synthase) together form the glutamine-dependent CPSase (GD-CPSase) (EC 6.3.5.5).
Similarity
Belongs to the glycosyl hydrolase 35 family.
In the central section; belongs to the metallo-dependent hydrolases superfamily. DHOase family. CAD subfamily.
In the central section; belongs to the metallo-dependent hydrolases superfamily. DHOase family. CAD subfamily.
Keywords
ATP-binding
Complete proteome
Cytoplasm
Glutamine amidotransferase
Hydrolase
Ligase
Magnesium
Manganese
Metal-binding
Multifunctional enzyme
Nucleotide-binding
Phosphoprotein
Pyrimidine biosynthesis
Reference proteome
Repeat
Transferase
Zinc
Feature
chain Beta-galactosidase
Uniprot
H9JBC1
A0A2A4K6E1
A0A194PFG8
A0A194RC64
A0A212FGU2
A0A2H1WN59
+ More
W4VRP3 A0A1Q3EWU3 A0A0P6IU92 B0WJ24 D6X207 A0A2J7PPL0 A0A0T6B8V4 A0A3F2YSP7 A0A182MTG8 A0A0L0CHK7 A0A182Q907 Q7QF47 A0A1Y1K525 A0A1I8QD31 A0A0A1XKF7 W8AXF4 A0A336KI92 T1PDM8 A0A1I8MPS8 A0A182YH98 A0A0K8U7R4 A0A084WPF6 A0A154PGP6 A0A1B6DTX5 A0A182JI15 A0A1W4XLI6 A0A195F8H4 A0A3F2Z211 A0A0A9XYC9 A0A1Y1K461 A0A151I8B4 A0A195BNQ3 A0A151J6A4 A0A151X4H0 A0A023F583 B4L1V5 E0V8W4 A0A1B0B005 B4M2B0 A0A1B0A3E5 B4JL50 A0A1W4VI53 A0A1B0FAD0 B3N0F3 A0A1A9W335 Q29HK5 B3NVK4 A0A1B6MQM2 B4MSD0 A0A1A9YDH3 X2JFG0 P05990 A0A3L8E3A1 E9G341 B4PXW0 A0A0P4X430 K7ITD2 A0A0P6HGF6 A0A0P5TIP9 A0A0P5RBU6 A0A0P5BE44 A0A162P1A7 A0A3R7M2Q2 A0A146MAP1 A0A0P4VY81 A0A1D2N6Q7 A0A0L7LMV3 A0A026WGC5 A0A0C9QL25 U3JAV8 A0A3N0Z596 A0A3Q1FF19 A0A3B4BMG0 A0A2D0PTD7 Q5XLV0 Q5EI67 A0A1S3J7E5 A0A3Q1FF01 A0A1W4YLN9 A0A0E3X9Y8 V9K783 W5NFY8 W5NFZ0 A0A2I4CN31 W5LFG9 A0A0F8C3Q0 A0A3P8ZEN2 A0A3P8ZF31 A0A3B1II39 A0A1S3S8T6 A0A3B4Z724 A0A087X8R3 A0A3Q0QVU3 A0A1S3S8T2
W4VRP3 A0A1Q3EWU3 A0A0P6IU92 B0WJ24 D6X207 A0A2J7PPL0 A0A0T6B8V4 A0A3F2YSP7 A0A182MTG8 A0A0L0CHK7 A0A182Q907 Q7QF47 A0A1Y1K525 A0A1I8QD31 A0A0A1XKF7 W8AXF4 A0A336KI92 T1PDM8 A0A1I8MPS8 A0A182YH98 A0A0K8U7R4 A0A084WPF6 A0A154PGP6 A0A1B6DTX5 A0A182JI15 A0A1W4XLI6 A0A195F8H4 A0A3F2Z211 A0A0A9XYC9 A0A1Y1K461 A0A151I8B4 A0A195BNQ3 A0A151J6A4 A0A151X4H0 A0A023F583 B4L1V5 E0V8W4 A0A1B0B005 B4M2B0 A0A1B0A3E5 B4JL50 A0A1W4VI53 A0A1B0FAD0 B3N0F3 A0A1A9W335 Q29HK5 B3NVK4 A0A1B6MQM2 B4MSD0 A0A1A9YDH3 X2JFG0 P05990 A0A3L8E3A1 E9G341 B4PXW0 A0A0P4X430 K7ITD2 A0A0P6HGF6 A0A0P5TIP9 A0A0P5RBU6 A0A0P5BE44 A0A162P1A7 A0A3R7M2Q2 A0A146MAP1 A0A0P4VY81 A0A1D2N6Q7 A0A0L7LMV3 A0A026WGC5 A0A0C9QL25 U3JAV8 A0A3N0Z596 A0A3Q1FF19 A0A3B4BMG0 A0A2D0PTD7 Q5XLV0 Q5EI67 A0A1S3J7E5 A0A3Q1FF01 A0A1W4YLN9 A0A0E3X9Y8 V9K783 W5NFY8 W5NFZ0 A0A2I4CN31 W5LFG9 A0A0F8C3Q0 A0A3P8ZEN2 A0A3P8ZF31 A0A3B1II39 A0A1S3S8T6 A0A3B4Z724 A0A087X8R3 A0A3Q0QVU3 A0A1S3S8T2
EC Number
3.2.1.23
Pubmed
19121390
26354079
22118469
26999592
18362917
19820115
+ More
26108605 12364791 14747013 17210077 28004739 25830018 24495485 25315136 25244985 24438588 25401762 25474469 17994087 20566863 18057021 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2884325 3023623 1329025 12537569 10080891 7932764 18327897 30249741 21292972 17550304 20075255 26823975 27289101 26227816 24508170 23594743 15256591 15937129 26383154 24402279 25329095 25835551 25069045
26108605 12364791 14747013 17210077 28004739 25830018 24495485 25315136 25244985 24438588 25401762 25474469 17994087 20566863 18057021 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2884325 3023623 1329025 12537569 10080891 7932764 18327897 30249741 21292972 17550304 20075255 26823975 27289101 26227816 24508170 23594743 15256591 15937129 26383154 24402279 25329095 25835551 25069045
EMBL
BABH01030910
NWSH01000130
PCG79242.1
KQ459606
KPI91444.1
KQ460398
+ More
KPJ15059.1 AGBW02008596 OWR52955.1 ODYU01009765 SOQ54416.1 GANO01000611 JAB59260.1 GFDL01015270 JAV19775.1 GDUN01000604 JAN95315.1 DS231955 EDS28904.1 KQ971371 EFA10197.1 NEVH01022660 PNF18278.1 LJIG01009100 KRT83756.1 APCN01001274 APCN01001275 APCN01001276 AXCM01002730 JRES01000391 KNC31702.1 AXCN02002019 AAAB01008846 EAA06526.5 GEZM01096981 JAV54456.1 GBXI01002488 JAD11804.1 GAMC01021069 JAB85486.1 UFQS01000523 UFQT01000523 SSX04574.1 SSX24937.1 KA646799 AFP61428.1 GDHF01029939 JAI22375.1 ATLV01025075 KE525369 KFB52100.1 KQ434890 KZC10488.1 GEDC01008180 JAS29118.1 KQ981744 KYN36492.1 GBHO01017797 JAG25807.1 GEZM01096996 JAV54415.1 KQ978377 KYM94554.1 KQ976438 KYM86863.1 KQ979878 KYN18700.1 KQ982554 KYQ55118.1 GBBI01002464 JAC16248.1 CH933810 EDW06758.1 DS234986 EEB09820.1 JXJN01006504 JXJN01006505 CH940651 EDW65814.1 KRF82430.1 CH916370 EDW00303.1 CCAG010017005 CH902640 EDV38357.1 KPU77376.1 KPU77377.1 CH379064 EAL31753.2 KRT06510.1 CH954180 EDV46396.1 GEBQ01001730 JAT38247.1 CH963851 EDW75019.1 AE014298 AHN59813.1 X04813 X03875 X03876 X03877 X03878 X03879 BT046159 M37783 AY089560 AF129814 S74010 QOIP01000001 RLU26933.1 GL732530 EFX86149.1 CM000162 EDX01946.1 GDIP01250655 JAI72746.1 AAZX01003296 AAZX01004114 GDIQ01020165 JAN74572.1 GDIP01131475 JAL72239.1 GDIQ01108117 JAL43609.1 GDIP01191108 JAJ32294.1 LRGB01000512 KZS18435.1 QCYY01002459 ROT70159.1 GDHC01001798 JAQ16831.1 GDRN01104231 JAI57956.1 LJIJ01000194 ODN00656.1 JTDY01000531 KOB76772.1 KK107260 EZA54099.1 GBYB01001247 JAG71014.1 BX511178 RJVU01008096 ROL53639.1 AY751464 AAU95538.1 AY880246 AAW80263.1 KP205549 AKC01947.1 JW861135 AFO93652.1 AHAT01008367 AHAT01008368 AHAT01008369 KQ041710 KKF23723.1 AYCK01020445
KPJ15059.1 AGBW02008596 OWR52955.1 ODYU01009765 SOQ54416.1 GANO01000611 JAB59260.1 GFDL01015270 JAV19775.1 GDUN01000604 JAN95315.1 DS231955 EDS28904.1 KQ971371 EFA10197.1 NEVH01022660 PNF18278.1 LJIG01009100 KRT83756.1 APCN01001274 APCN01001275 APCN01001276 AXCM01002730 JRES01000391 KNC31702.1 AXCN02002019 AAAB01008846 EAA06526.5 GEZM01096981 JAV54456.1 GBXI01002488 JAD11804.1 GAMC01021069 JAB85486.1 UFQS01000523 UFQT01000523 SSX04574.1 SSX24937.1 KA646799 AFP61428.1 GDHF01029939 JAI22375.1 ATLV01025075 KE525369 KFB52100.1 KQ434890 KZC10488.1 GEDC01008180 JAS29118.1 KQ981744 KYN36492.1 GBHO01017797 JAG25807.1 GEZM01096996 JAV54415.1 KQ978377 KYM94554.1 KQ976438 KYM86863.1 KQ979878 KYN18700.1 KQ982554 KYQ55118.1 GBBI01002464 JAC16248.1 CH933810 EDW06758.1 DS234986 EEB09820.1 JXJN01006504 JXJN01006505 CH940651 EDW65814.1 KRF82430.1 CH916370 EDW00303.1 CCAG010017005 CH902640 EDV38357.1 KPU77376.1 KPU77377.1 CH379064 EAL31753.2 KRT06510.1 CH954180 EDV46396.1 GEBQ01001730 JAT38247.1 CH963851 EDW75019.1 AE014298 AHN59813.1 X04813 X03875 X03876 X03877 X03878 X03879 BT046159 M37783 AY089560 AF129814 S74010 QOIP01000001 RLU26933.1 GL732530 EFX86149.1 CM000162 EDX01946.1 GDIP01250655 JAI72746.1 AAZX01003296 AAZX01004114 GDIQ01020165 JAN74572.1 GDIP01131475 JAL72239.1 GDIQ01108117 JAL43609.1 GDIP01191108 JAJ32294.1 LRGB01000512 KZS18435.1 QCYY01002459 ROT70159.1 GDHC01001798 JAQ16831.1 GDRN01104231 JAI57956.1 LJIJ01000194 ODN00656.1 JTDY01000531 KOB76772.1 KK107260 EZA54099.1 GBYB01001247 JAG71014.1 BX511178 RJVU01008096 ROL53639.1 AY751464 AAU95538.1 AY880246 AAW80263.1 KP205549 AKC01947.1 JW861135 AFO93652.1 AHAT01008367 AHAT01008368 AHAT01008369 KQ041710 KKF23723.1 AYCK01020445
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000002320
+ More
UP000007266 UP000235965 UP000075840 UP000075883 UP000037069 UP000075886 UP000007062 UP000095300 UP000095301 UP000076408 UP000030765 UP000076502 UP000075880 UP000192223 UP000078541 UP000076407 UP000078542 UP000078540 UP000078492 UP000075809 UP000009192 UP000009046 UP000092460 UP000008792 UP000092445 UP000001070 UP000192221 UP000092444 UP000007801 UP000091820 UP000001819 UP000008711 UP000007798 UP000092443 UP000000803 UP000279307 UP000000305 UP000002282 UP000002358 UP000076858 UP000283509 UP000094527 UP000037510 UP000053097 UP000000437 UP000257200 UP000261440 UP000221080 UP000085678 UP000192224 UP000018468 UP000192220 UP000018467 UP000265140 UP000087266 UP000261400 UP000028760 UP000261340
UP000007266 UP000235965 UP000075840 UP000075883 UP000037069 UP000075886 UP000007062 UP000095300 UP000095301 UP000076408 UP000030765 UP000076502 UP000075880 UP000192223 UP000078541 UP000076407 UP000078542 UP000078540 UP000078492 UP000075809 UP000009192 UP000009046 UP000092460 UP000008792 UP000092445 UP000001070 UP000192221 UP000092444 UP000007801 UP000091820 UP000001819 UP000008711 UP000007798 UP000092443 UP000000803 UP000279307 UP000000305 UP000002282 UP000002358 UP000076858 UP000283509 UP000094527 UP000037510 UP000053097 UP000000437 UP000257200 UP000261440 UP000221080 UP000085678 UP000192224 UP000018468 UP000192220 UP000018467 UP000265140 UP000087266 UP000261400 UP000028760 UP000261340
Pfam
Interpro
IPR002474
CarbamoylP_synth_ssu_N
+ More
IPR006275 CarbamoylP_synth_lsu
IPR017926 GATASE
IPR036480 CarbP_synth_ssu_N_sf
IPR005480 CarbamoylP_synth_lsu_oligo
IPR006132 Asp/Orn_carbamoyltranf_P-bd
IPR036901 Asp/Orn_carbamoylTrfase_sf
IPR005483 CbamoylP_synth_lsu_CPSase_dom
IPR011059 Metal-dep_hydrolase_composite
IPR013815 ATP_grasp_subdomain_1
IPR036914 MGS-like_dom_sf
IPR006680 Amidohydro-rel
IPR006130 Asp/Orn_carbamoylTrfase
IPR005479 CbamoylP_synth_lsu-like_ATP-bd
IPR011607 MGS-like_dom
IPR035686 CPSase_GATase1
IPR016185 PreATP-grasp_dom_sf
IPR006131 Asp_carbamoyltransf_Asp/Orn-bd
IPR032466 Metal_Hydrolase
IPR011761 ATP-grasp
IPR002082 Asp_carbamoyltransf
IPR036897 CarbamoylP_synth_lsu_oligo_sf
IPR002195 Dihydroorotase_CS
IPR029062 Class_I_gatase-like
IPR006274 CarbamoylP_synth_ssu
IPR001944 Glycoside_Hdrlase_35
IPR025300 BetaGal_jelly_roll_dom
IPR008979 Galactose-bd-like_sf
IPR017853 Glycoside_hydrolase_SF
IPR019801 Glyco_hydro_35_CS
IPR031330 Gly_Hdrlase_35_cat
IPR018253 DnaJ_domain_CS
IPR036869 J_dom_sf
IPR001623 DnaJ_domain
IPR006275 CarbamoylP_synth_lsu
IPR017926 GATASE
IPR036480 CarbP_synth_ssu_N_sf
IPR005480 CarbamoylP_synth_lsu_oligo
IPR006132 Asp/Orn_carbamoyltranf_P-bd
IPR036901 Asp/Orn_carbamoylTrfase_sf
IPR005483 CbamoylP_synth_lsu_CPSase_dom
IPR011059 Metal-dep_hydrolase_composite
IPR013815 ATP_grasp_subdomain_1
IPR036914 MGS-like_dom_sf
IPR006680 Amidohydro-rel
IPR006130 Asp/Orn_carbamoylTrfase
IPR005479 CbamoylP_synth_lsu-like_ATP-bd
IPR011607 MGS-like_dom
IPR035686 CPSase_GATase1
IPR016185 PreATP-grasp_dom_sf
IPR006131 Asp_carbamoyltransf_Asp/Orn-bd
IPR032466 Metal_Hydrolase
IPR011761 ATP-grasp
IPR002082 Asp_carbamoyltransf
IPR036897 CarbamoylP_synth_lsu_oligo_sf
IPR002195 Dihydroorotase_CS
IPR029062 Class_I_gatase-like
IPR006274 CarbamoylP_synth_ssu
IPR001944 Glycoside_Hdrlase_35
IPR025300 BetaGal_jelly_roll_dom
IPR008979 Galactose-bd-like_sf
IPR017853 Glycoside_hydrolase_SF
IPR019801 Glyco_hydro_35_CS
IPR031330 Gly_Hdrlase_35_cat
IPR018253 DnaJ_domain_CS
IPR036869 J_dom_sf
IPR001623 DnaJ_domain
SUPFAM
Gene 3D
ProteinModelPortal
H9JBC1
A0A2A4K6E1
A0A194PFG8
A0A194RC64
A0A212FGU2
A0A2H1WN59
+ More
W4VRP3 A0A1Q3EWU3 A0A0P6IU92 B0WJ24 D6X207 A0A2J7PPL0 A0A0T6B8V4 A0A3F2YSP7 A0A182MTG8 A0A0L0CHK7 A0A182Q907 Q7QF47 A0A1Y1K525 A0A1I8QD31 A0A0A1XKF7 W8AXF4 A0A336KI92 T1PDM8 A0A1I8MPS8 A0A182YH98 A0A0K8U7R4 A0A084WPF6 A0A154PGP6 A0A1B6DTX5 A0A182JI15 A0A1W4XLI6 A0A195F8H4 A0A3F2Z211 A0A0A9XYC9 A0A1Y1K461 A0A151I8B4 A0A195BNQ3 A0A151J6A4 A0A151X4H0 A0A023F583 B4L1V5 E0V8W4 A0A1B0B005 B4M2B0 A0A1B0A3E5 B4JL50 A0A1W4VI53 A0A1B0FAD0 B3N0F3 A0A1A9W335 Q29HK5 B3NVK4 A0A1B6MQM2 B4MSD0 A0A1A9YDH3 X2JFG0 P05990 A0A3L8E3A1 E9G341 B4PXW0 A0A0P4X430 K7ITD2 A0A0P6HGF6 A0A0P5TIP9 A0A0P5RBU6 A0A0P5BE44 A0A162P1A7 A0A3R7M2Q2 A0A146MAP1 A0A0P4VY81 A0A1D2N6Q7 A0A0L7LMV3 A0A026WGC5 A0A0C9QL25 U3JAV8 A0A3N0Z596 A0A3Q1FF19 A0A3B4BMG0 A0A2D0PTD7 Q5XLV0 Q5EI67 A0A1S3J7E5 A0A3Q1FF01 A0A1W4YLN9 A0A0E3X9Y8 V9K783 W5NFY8 W5NFZ0 A0A2I4CN31 W5LFG9 A0A0F8C3Q0 A0A3P8ZEN2 A0A3P8ZF31 A0A3B1II39 A0A1S3S8T6 A0A3B4Z724 A0A087X8R3 A0A3Q0QVU3 A0A1S3S8T2
W4VRP3 A0A1Q3EWU3 A0A0P6IU92 B0WJ24 D6X207 A0A2J7PPL0 A0A0T6B8V4 A0A3F2YSP7 A0A182MTG8 A0A0L0CHK7 A0A182Q907 Q7QF47 A0A1Y1K525 A0A1I8QD31 A0A0A1XKF7 W8AXF4 A0A336KI92 T1PDM8 A0A1I8MPS8 A0A182YH98 A0A0K8U7R4 A0A084WPF6 A0A154PGP6 A0A1B6DTX5 A0A182JI15 A0A1W4XLI6 A0A195F8H4 A0A3F2Z211 A0A0A9XYC9 A0A1Y1K461 A0A151I8B4 A0A195BNQ3 A0A151J6A4 A0A151X4H0 A0A023F583 B4L1V5 E0V8W4 A0A1B0B005 B4M2B0 A0A1B0A3E5 B4JL50 A0A1W4VI53 A0A1B0FAD0 B3N0F3 A0A1A9W335 Q29HK5 B3NVK4 A0A1B6MQM2 B4MSD0 A0A1A9YDH3 X2JFG0 P05990 A0A3L8E3A1 E9G341 B4PXW0 A0A0P4X430 K7ITD2 A0A0P6HGF6 A0A0P5TIP9 A0A0P5RBU6 A0A0P5BE44 A0A162P1A7 A0A3R7M2Q2 A0A146MAP1 A0A0P4VY81 A0A1D2N6Q7 A0A0L7LMV3 A0A026WGC5 A0A0C9QL25 U3JAV8 A0A3N0Z596 A0A3Q1FF19 A0A3B4BMG0 A0A2D0PTD7 Q5XLV0 Q5EI67 A0A1S3J7E5 A0A3Q1FF01 A0A1W4YLN9 A0A0E3X9Y8 V9K783 W5NFY8 W5NFZ0 A0A2I4CN31 W5LFG9 A0A0F8C3Q0 A0A3P8ZEN2 A0A3P8ZF31 A0A3B1II39 A0A1S3S8T6 A0A3B4Z724 A0A087X8R3 A0A3Q0QVU3 A0A1S3S8T2
PDB
5DOU
E-value=0,
Score=3629
Ontologies
KEGG
GO
GO:0046872
GO:0006541
GO:0004070
GO:0006207
GO:0005524
GO:0016597
GO:0016812
GO:0004088
GO:0005975
GO:0004565
GO:0006228
GO:0006807
GO:0019240
GO:0005829
GO:0005737
GO:0004151
GO:0004087
GO:0070406
GO:0044205
GO:0016021
GO:0006520
GO:0060041
GO:0048812
GO:1900746
GO:0021636
GO:0030947
GO:0021610
GO:0008593
GO:0021644
GO:0021615
GO:0001946
GO:0016743
GO:0016810
GO:0016787
Topology
Subcellular location
Cytoplasm
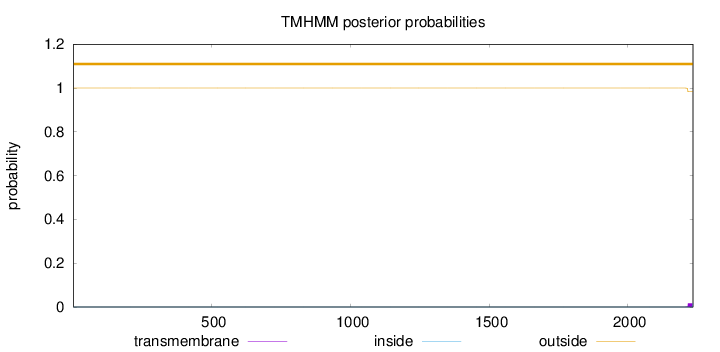
Length:
2235
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.30998
Exp number, first 60 AAs:
0.000900000000000001
Total prob of N-in:
0.00014
outside
1 - 2235
Population Genetic Test Statistics
Pi
226.115299
Theta
215.188199
Tajima's D
0.479401
CLR
0.165877
CSRT
0.512074396280186
Interpretation
Uncertain
