Gene
KWMTBOMO05724
Pre Gene Modal
BGIBMGA006869
Annotation
PREDICTED:_CKLF-like_MARVEL_transmembrane_domain-containing_protein_4_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.837
Sequence
CDS
ATGATGGCACCCGAGACAGTAGTAACTGTGGACCATAACAAACCTGCTGCGGCACCTCCTCCACAAGGCGGTGCACTCGACTGGATTAAAATCAACGTCGACTACTTCAAAACCCCACCTGGCTTGCTCAAAATAATTCAGTTGATTCTCGGCATACTATGTATGGGGTTAGGCTCTCCGTGGTACTCGACGTGGTTCGTCTTCGTCGCAGTCACCAGCTTCATCACAACCCTCATGTGGAGTTTCGTATACTTACTCAGCATCAGGGAGGCTCTTAAGCTGCCCATTAATTGGGTTCTTTCTGAATTAATTAGTACAGCACTGGAGACAGTAATGTATCTGTTGGCGTTCATAGTTCTGTTTGCTGCGACGGGAACTTATTATGGACCAAGATACGGAAGCAATGTTGCAGCTGCCGTTTTCGGTATCTTTAACACCATCGCATACGGAGCCAGTTCGTTTCTGCTGTTTAGAGAACACAGGGCCTCCTCCACGGCGACACAAGCAGCGTGA
Protein
MMAPETVVTVDHNKPAAAPPPQGGALDWIKINVDYFKTPPGLLKIIQLILGILCMGLGSPWYSTWFVFVAVTSFITTLMWSFVYLLSIREALKLPINWVLSELISTALETVMYLLAFIVLFAATGTYYGPRYGSNVAAAVFGIFNTIAYGASSFLLFREHRASSTATQAA
Summary
Uniprot
A0A2A4K5Q4
A0A1E1WC62
I4DJ20
I4DML3
A0A212F6V5
U5EQZ4
+ More
A0A023EHM6 A0A1L8E3H6 A0A182GX90 A0A1W4XVG6 A0A1Q3FZF5 A0A0A9YY87 A0A2M4AZC4 A0A1I8PRS7 A0A2M4CRS3 A0A2M4AZB7 A0A2M4C0T2 A0A139WCB4 A0A2M3Z9C9 A0A1B6H739 A0A182FMN8 A0A1Q3FZC0 T1D6H9 A0A1B6MKG5 V9IJW8 A0A182YH95 A0A2S2NMC7 A0A0N7Z8N2 A0A151J642 A0A2H8TIV5 V5G4A3 A0A151WKX8 A0A1B6GQT7 J9K1K8 A0A158ND55 A0A0C9RUN1 A0A2S2Q785 W8CCG7 A0A182XBW8 A0A182IAL6 A0A182TIV0 Q7QF43 A0A034WF78 A0A0K8W8I5 A0A182QBM2 A0A1L8EGQ6 A0A182N1F3 J7H5H7 B3MQD3 A0A182ILB7 A0A1W4V2T8 Q29H35 D0QWF0 A0A3B0K5L1 A0A1J1IIH2 B4N1J8 A0A0L0CB08 A0A0P4WDE6 B4PXZ6 Q9VZ71 B3NVE7 B4IDT9 B4R7M7 J7H8M2 K7ITC9 A0A1D2N5U7
A0A023EHM6 A0A1L8E3H6 A0A182GX90 A0A1W4XVG6 A0A1Q3FZF5 A0A0A9YY87 A0A2M4AZC4 A0A1I8PRS7 A0A2M4CRS3 A0A2M4AZB7 A0A2M4C0T2 A0A139WCB4 A0A2M3Z9C9 A0A1B6H739 A0A182FMN8 A0A1Q3FZC0 T1D6H9 A0A1B6MKG5 V9IJW8 A0A182YH95 A0A2S2NMC7 A0A0N7Z8N2 A0A151J642 A0A2H8TIV5 V5G4A3 A0A151WKX8 A0A1B6GQT7 J9K1K8 A0A158ND55 A0A0C9RUN1 A0A2S2Q785 W8CCG7 A0A182XBW8 A0A182IAL6 A0A182TIV0 Q7QF43 A0A034WF78 A0A0K8W8I5 A0A182QBM2 A0A1L8EGQ6 A0A182N1F3 J7H5H7 B3MQD3 A0A182ILB7 A0A1W4V2T8 Q29H35 D0QWF0 A0A3B0K5L1 A0A1J1IIH2 B4N1J8 A0A0L0CB08 A0A0P4WDE6 B4PXZ6 Q9VZ71 B3NVE7 B4IDT9 B4R7M7 J7H8M2 K7ITC9 A0A1D2N5U7
Pubmed
22651552
22118469
24945155
26483478
25401762
26823975
+ More
18362917 19820115 24330624 25244985 27129103 21347285 24495485 12364791 14747013 17210077 25348373 22766100 17994087 15632085 19859648 26108605 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 27289101
18362917 19820115 24330624 25244985 27129103 21347285 24495485 12364791 14747013 17210077 25348373 22766100 17994087 15632085 19859648 26108605 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 27289101
EMBL
NWSH01000130
PCG79248.1
GDQN01006505
JAT84549.1
AK401288
BAM17910.1
+ More
AK402531 BAM19153.1 AGBW02009960 OWR49476.1 GANO01003985 JAB55886.1 GAPW01005056 GAPW01005055 JAC08542.1 GFDF01001029 JAV13055.1 JXUM01094931 JXUM01094932 KQ564162 KXJ72653.1 GFDL01002139 JAV32906.1 GBHO01005612 GBHO01005611 GBRD01002140 GBRD01002139 GBRD01002138 GDHC01004871 GDHC01003252 JAG37992.1 JAG37993.1 JAG63681.1 JAQ13758.1 JAQ15377.1 GGFK01012784 MBW46105.1 GGFL01003797 MBW67975.1 GGFK01012741 MBW46062.1 GGFJ01009754 MBW58895.1 KQ971371 KYB25544.1 GGFM01004287 MBW25038.1 GECU01037159 GECU01013530 JAS70547.1 JAS94176.1 GFDL01002147 JAV32898.1 GALA01000048 JAA94804.1 GEBQ01003529 JAT36448.1 JR048870 AEY60786.1 GGMR01005711 MBY18330.1 GDKW01002875 JAI53720.1 KQ979878 KYN18694.1 GFXV01002231 MBW14036.1 GALX01003601 JAB64865.1 KQ982987 KYQ48532.1 GECZ01032357 GECZ01024411 GECZ01022050 GECZ01019093 GECZ01014852 GECZ01014670 GECZ01004975 JAS37412.1 JAS45358.1 JAS47719.1 JAS50676.1 JAS54917.1 JAS55099.1 JAS64794.1 ABLF02035431 ADTU01012401 ADTU01012402 ADTU01012403 GBYB01012565 JAG82332.1 GGMS01004396 MBY73599.1 GAMC01003727 GAMC01003726 JAC02830.1 APCN01001271 AAAB01008846 EAA06210.4 GAKP01006167 JAC52785.1 GDHF01030729 GDHF01029948 GDHF01020153 GDHF01005134 GDHF01001070 GDHF01000976 GDHF01000343 JAI21585.1 JAI22366.1 JAI32161.1 JAI47180.1 JAI51244.1 JAI51338.1 JAI51971.1 AXCN02002019 GFDG01000951 JAV17848.1 JX012239 AFP92127.1 CH902621 EDV44559.1 CH379064 EAL31924.1 FJ821026 ACN94643.1 OUUW01000021 SPP89415.1 CVRI01000054 CRL00000.1 CH963925 EDW78237.1 JRES01000755 KNC28659.1 GDRN01071745 JAI63643.1 CM000162 EDX02968.1 AE014298 AY069765 AAF47955.1 AAL39910.1 ABW09385.1 CH954180 EDV46135.1 CH480830 EDW45747.1 CM000366 EDX17586.1 JX012238 AFP92126.1 AAZX01003296 LJIJ01000194 ODN00649.1
AK402531 BAM19153.1 AGBW02009960 OWR49476.1 GANO01003985 JAB55886.1 GAPW01005056 GAPW01005055 JAC08542.1 GFDF01001029 JAV13055.1 JXUM01094931 JXUM01094932 KQ564162 KXJ72653.1 GFDL01002139 JAV32906.1 GBHO01005612 GBHO01005611 GBRD01002140 GBRD01002139 GBRD01002138 GDHC01004871 GDHC01003252 JAG37992.1 JAG37993.1 JAG63681.1 JAQ13758.1 JAQ15377.1 GGFK01012784 MBW46105.1 GGFL01003797 MBW67975.1 GGFK01012741 MBW46062.1 GGFJ01009754 MBW58895.1 KQ971371 KYB25544.1 GGFM01004287 MBW25038.1 GECU01037159 GECU01013530 JAS70547.1 JAS94176.1 GFDL01002147 JAV32898.1 GALA01000048 JAA94804.1 GEBQ01003529 JAT36448.1 JR048870 AEY60786.1 GGMR01005711 MBY18330.1 GDKW01002875 JAI53720.1 KQ979878 KYN18694.1 GFXV01002231 MBW14036.1 GALX01003601 JAB64865.1 KQ982987 KYQ48532.1 GECZ01032357 GECZ01024411 GECZ01022050 GECZ01019093 GECZ01014852 GECZ01014670 GECZ01004975 JAS37412.1 JAS45358.1 JAS47719.1 JAS50676.1 JAS54917.1 JAS55099.1 JAS64794.1 ABLF02035431 ADTU01012401 ADTU01012402 ADTU01012403 GBYB01012565 JAG82332.1 GGMS01004396 MBY73599.1 GAMC01003727 GAMC01003726 JAC02830.1 APCN01001271 AAAB01008846 EAA06210.4 GAKP01006167 JAC52785.1 GDHF01030729 GDHF01029948 GDHF01020153 GDHF01005134 GDHF01001070 GDHF01000976 GDHF01000343 JAI21585.1 JAI22366.1 JAI32161.1 JAI47180.1 JAI51244.1 JAI51338.1 JAI51971.1 AXCN02002019 GFDG01000951 JAV17848.1 JX012239 AFP92127.1 CH902621 EDV44559.1 CH379064 EAL31924.1 FJ821026 ACN94643.1 OUUW01000021 SPP89415.1 CVRI01000054 CRL00000.1 CH963925 EDW78237.1 JRES01000755 KNC28659.1 GDRN01071745 JAI63643.1 CM000162 EDX02968.1 AE014298 AY069765 AAF47955.1 AAL39910.1 ABW09385.1 CH954180 EDV46135.1 CH480830 EDW45747.1 CM000366 EDX17586.1 JX012238 AFP92126.1 AAZX01003296 LJIJ01000194 ODN00649.1
Proteomes
UP000218220
UP000007151
UP000069940
UP000249989
UP000192223
UP000095300
+ More
UP000007266 UP000069272 UP000076408 UP000078492 UP000075809 UP000007819 UP000005205 UP000076407 UP000075840 UP000075902 UP000007062 UP000075886 UP000075884 UP000007801 UP000075880 UP000192221 UP000001819 UP000268350 UP000183832 UP000007798 UP000037069 UP000002282 UP000000803 UP000008711 UP000001292 UP000000304 UP000002358 UP000094527
UP000007266 UP000069272 UP000076408 UP000078492 UP000075809 UP000007819 UP000005205 UP000076407 UP000075840 UP000075902 UP000007062 UP000075886 UP000075884 UP000007801 UP000075880 UP000192221 UP000001819 UP000268350 UP000183832 UP000007798 UP000037069 UP000002282 UP000000803 UP000008711 UP000001292 UP000000304 UP000002358 UP000094527
PRIDE
Pfam
PF01284 MARVEL
ProteinModelPortal
A0A2A4K5Q4
A0A1E1WC62
I4DJ20
I4DML3
A0A212F6V5
U5EQZ4
+ More
A0A023EHM6 A0A1L8E3H6 A0A182GX90 A0A1W4XVG6 A0A1Q3FZF5 A0A0A9YY87 A0A2M4AZC4 A0A1I8PRS7 A0A2M4CRS3 A0A2M4AZB7 A0A2M4C0T2 A0A139WCB4 A0A2M3Z9C9 A0A1B6H739 A0A182FMN8 A0A1Q3FZC0 T1D6H9 A0A1B6MKG5 V9IJW8 A0A182YH95 A0A2S2NMC7 A0A0N7Z8N2 A0A151J642 A0A2H8TIV5 V5G4A3 A0A151WKX8 A0A1B6GQT7 J9K1K8 A0A158ND55 A0A0C9RUN1 A0A2S2Q785 W8CCG7 A0A182XBW8 A0A182IAL6 A0A182TIV0 Q7QF43 A0A034WF78 A0A0K8W8I5 A0A182QBM2 A0A1L8EGQ6 A0A182N1F3 J7H5H7 B3MQD3 A0A182ILB7 A0A1W4V2T8 Q29H35 D0QWF0 A0A3B0K5L1 A0A1J1IIH2 B4N1J8 A0A0L0CB08 A0A0P4WDE6 B4PXZ6 Q9VZ71 B3NVE7 B4IDT9 B4R7M7 J7H8M2 K7ITC9 A0A1D2N5U7
A0A023EHM6 A0A1L8E3H6 A0A182GX90 A0A1W4XVG6 A0A1Q3FZF5 A0A0A9YY87 A0A2M4AZC4 A0A1I8PRS7 A0A2M4CRS3 A0A2M4AZB7 A0A2M4C0T2 A0A139WCB4 A0A2M3Z9C9 A0A1B6H739 A0A182FMN8 A0A1Q3FZC0 T1D6H9 A0A1B6MKG5 V9IJW8 A0A182YH95 A0A2S2NMC7 A0A0N7Z8N2 A0A151J642 A0A2H8TIV5 V5G4A3 A0A151WKX8 A0A1B6GQT7 J9K1K8 A0A158ND55 A0A0C9RUN1 A0A2S2Q785 W8CCG7 A0A182XBW8 A0A182IAL6 A0A182TIV0 Q7QF43 A0A034WF78 A0A0K8W8I5 A0A182QBM2 A0A1L8EGQ6 A0A182N1F3 J7H5H7 B3MQD3 A0A182ILB7 A0A1W4V2T8 Q29H35 D0QWF0 A0A3B0K5L1 A0A1J1IIH2 B4N1J8 A0A0L0CB08 A0A0P4WDE6 B4PXZ6 Q9VZ71 B3NVE7 B4IDT9 B4R7M7 J7H8M2 K7ITC9 A0A1D2N5U7
Ontologies
GO
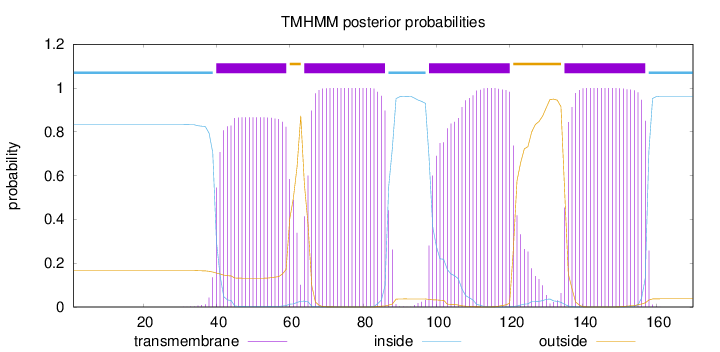
Topology
Length:
170
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
85.9330100000001
Exp number, first 60 AAs:
17.41406
Total prob of N-in:
0.83349
POSSIBLE N-term signal
sequence
inside
1 - 39
TMhelix
40 - 59
outside
60 - 63
TMhelix
64 - 86
inside
87 - 97
TMhelix
98 - 120
outside
121 - 134
TMhelix
135 - 157
inside
158 - 170
Population Genetic Test Statistics
Pi
234.626255
Theta
194.387288
Tajima's D
0.782391
CLR
0.356268
CSRT
0.595970201489926
Interpretation
Uncertain
