Pre Gene Modal
BGIBMGA006866
Annotation
PREDICTED:_uncharacterized_protein_LOC106131802_isoform_X4_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.027 Mitochondrial Reliability : 1.044 Nuclear Reliability : 1.653
Sequence
CDS
ATGACTGAACTGGAGACAAGGCAAGATGAACTCTTGAACAAGCTAGAAATACTATACGAGCGGATAAAGACAATTAGCTCTCAATGTAAGCTTGACAATATGAGTGCTAATATTCAGTCCTCAAATGCCTCAAAAGTCGAATCTGTAATTACACCAGAGGAAGTTGTCTTGGTGCTGAGTCCTGATTCATTGCCTTGGTATTTGAATATCATTTTGAAAAAATCCAATTTACCAATACATACAACTTGTCATGTCCATTCATCAGTGCCAAGTGAGAAATTAGCAAAAATAAAGGCTTTTACTCAAAAACTTAAAACTTCTGAAAACCCTAAAGTAAACCTAAGGCTTATTTTTAAAGCTGCAGCAGATTCAGAACTGAAATTGTCAGCATTATCAACACCAATCTTAGGAAATGTCAACATCCTAAGATATCTGTCATTAGCCTACCCTACCATAATTCCTTATGACCATAATGATCACATAGTTGATAATCTATTAGACATTTGCCACGTTTTGGAGAGGACATCAGAAAAAAATAAAGAAGCAGTTGTAAATAGGCTTTTCGCTCACTATAAAACTTGGATTTATGGTGACAAATTTTCTGTGGTAGACTTGGCAGCATACAACTTAATTAAGCAGTGGAGAAATACACCTAAGTTTGTCTCTAAAGTGTGGTTTGATAAATGTGAAAAATTATGTTCTTAA
Protein
MTELETRQDELLNKLEILYERIKTISSQCKLDNMSANIQSSNASKVESVITPEEVVLVLSPDSLPWYLNIILKKSNLPIHTTCHVHSSVPSEKLAKIKAFTQKLKTSENPKVNLRLIFKAAADSELKLSALSTPILGNVNILRYLSLAYPTIIPYDHNDHIVDNLLDICHVLERTSEKNKEAVVNRLFAHYKTWIYGDKFSVVDLAAYNLIKQWRNTPKFVSKVWFDKCEKLCS
Summary
Uniprot
H9JBH1
A0A2A4JAK5
A0A2A4J9T5
A0A2A4JB18
A0A2H1W975
A0A1E1WLK2
+ More
A0A1E1VXK7 A0A1E1WJ44 A0A1E1WB24 A0A0L7LIS2 A0A212F6X0 S4PPU5 A0A194PK35 A0A194RBU6 V5IA46 V5GXW2 A0A139WC74 A0A182WLT8 D6X1S7 A0A182Q9I0 A0A1B6M7B0 A0A1B6L3P0 A0A1B6MTX9 A0A182PVJ3 A0A182MCG9 Q7Q7A3 A0A182LJT6 A0A182HM92 A0A182XCW3 U4UCQ8 A0A1B6GU67 J3JWN3 A0A182RC29 A0A182TXA8 A0A182VBV3 N6T5H5 A0A182JV20 A0A1B6J935 A0A1B6IXX8 A0A1B6JPP6 A0A067R539 A0A182GH13 A0A0K8TUE9 R4G7X1 A0A0K8SMG8 A0A0A9YLU8 A0A2P8Y696 A0A1B6JCC8 A0A182Y0R2 A0A182J9X7 A0A2M3Z7U1 A0A069DYJ2 A0A2M4AHK6 A0A2M4BWW4 A0A084WDB7 A0A0M8ZUS9 A0A2M3Z958 A0A2M4BUH2 A0A2M4BUM1 E2BIY9 A0A182ST72 A0A182FHP3 Q174X6 A0A1S4FEE5 E2AE30 A0A2M4CGS7 A0A1Q3F070 A0A0V0G6A9 A0A2M4CGS1 W8BEE4 A0A182NMD4 W8BPB6 A0A1Y1LME9 A0A2M3Z7X0 A0A293MVX7 A0A1Y1LKT5 W5JNE3 A0A1Q3F2Q5 K7IT22 A0A2A3EPM9 A0A1I8ND92 T1PEX8 A0A1B6DMX2 A0A1B6CP86 A0A2R7VQI7 A0A1I8ND98 U4U0P6 B4LIJ0 A0A0Q9WV42 E9IWI1 A0A1J1HZK0 A0A2G8LE52 B0WXK6 A0A088ACJ5 A0A2J7RD15 A0A1I8Q993 A0A151WHZ0
A0A1E1VXK7 A0A1E1WJ44 A0A1E1WB24 A0A0L7LIS2 A0A212F6X0 S4PPU5 A0A194PK35 A0A194RBU6 V5IA46 V5GXW2 A0A139WC74 A0A182WLT8 D6X1S7 A0A182Q9I0 A0A1B6M7B0 A0A1B6L3P0 A0A1B6MTX9 A0A182PVJ3 A0A182MCG9 Q7Q7A3 A0A182LJT6 A0A182HM92 A0A182XCW3 U4UCQ8 A0A1B6GU67 J3JWN3 A0A182RC29 A0A182TXA8 A0A182VBV3 N6T5H5 A0A182JV20 A0A1B6J935 A0A1B6IXX8 A0A1B6JPP6 A0A067R539 A0A182GH13 A0A0K8TUE9 R4G7X1 A0A0K8SMG8 A0A0A9YLU8 A0A2P8Y696 A0A1B6JCC8 A0A182Y0R2 A0A182J9X7 A0A2M3Z7U1 A0A069DYJ2 A0A2M4AHK6 A0A2M4BWW4 A0A084WDB7 A0A0M8ZUS9 A0A2M3Z958 A0A2M4BUH2 A0A2M4BUM1 E2BIY9 A0A182ST72 A0A182FHP3 Q174X6 A0A1S4FEE5 E2AE30 A0A2M4CGS7 A0A1Q3F070 A0A0V0G6A9 A0A2M4CGS1 W8BEE4 A0A182NMD4 W8BPB6 A0A1Y1LME9 A0A2M3Z7X0 A0A293MVX7 A0A1Y1LKT5 W5JNE3 A0A1Q3F2Q5 K7IT22 A0A2A3EPM9 A0A1I8ND92 T1PEX8 A0A1B6DMX2 A0A1B6CP86 A0A2R7VQI7 A0A1I8ND98 U4U0P6 B4LIJ0 A0A0Q9WV42 E9IWI1 A0A1J1HZK0 A0A2G8LE52 B0WXK6 A0A088ACJ5 A0A2J7RD15 A0A1I8Q993 A0A151WHZ0
Pubmed
EMBL
BABH01030903
NWSH01002220
PCG68889.1
PCG68887.1
PCG68888.1
ODYU01007140
+ More
SOQ49649.1 GDQN01003165 GDQN01000896 JAT87889.1 JAT90158.1 GDQN01011586 JAT79468.1 GDQN01004039 JAT87015.1 GDQN01006909 JAT84145.1 JTDY01001022 KOB75106.1 AGBW02009960 OWR49480.1 GAIX01000007 JAA92553.1 KQ459606 KPI91455.1 KQ460398 KPJ15072.1 GALX01001820 JAB66646.1 GALX01001819 JAB66647.1 KQ971371 KYB25520.1 EFA09968.1 AXCN02000934 GEBQ01008172 JAT31805.1 GEBQ01021625 JAT18352.1 GEBQ01000597 JAT39380.1 AXCM01007726 AAAB01008960 EAA10870.4 APCN01004380 KB632204 ERL90108.1 GECZ01003808 JAS65961.1 BT127651 AEE62613.1 APGK01043377 APGK01043378 KB741014 ENN75454.1 GECU01011989 JAS95717.1 GECU01015943 JAS91763.1 GECU01006492 JAT01215.1 KK852687 KDR18392.1 JXUM01062867 KQ562219 KXJ76368.1 GDAI01000038 JAI17565.1 GAHY01001982 JAA75528.1 GBRD01011318 JAG54506.1 GBHO01011531 JAG32073.1 PYGN01000877 PSN39773.1 GECU01010790 JAS96916.1 GGFM01003820 MBW24571.1 GBGD01002295 JAC86594.1 GGFK01006939 MBW40260.1 GGFJ01008382 MBW57523.1 ATLV01022985 KE525339 KFB48211.1 KQ435840 KOX71397.1 GGFM01004269 MBW25020.1 GGFJ01007267 MBW56408.1 GGFJ01007317 MBW56458.1 GL448543 EFN84334.1 CH477404 EAT41655.1 EAT41656.1 GL438827 EFN68263.1 GGFL01000336 MBW64514.1 GFDL01014101 JAV20944.1 GECL01002597 JAP03527.1 GGFL01000335 MBW64513.1 GAMC01011192 JAB95363.1 GAMC01011194 JAB95361.1 GEZM01054604 JAV73530.1 GGFM01003863 MBW24614.1 GFWV01018742 MAA43470.1 GEZM01054603 JAV73531.1 ADMH02001054 ETN64294.1 GFDL01013205 JAV21840.1 AAZX01003332 KZ288204 PBC33222.1 KA646705 AFP61334.1 GEDC01010289 JAS27009.1 GEDC01022114 JAS15184.1 KK854028 PTY09784.1 KB630474 ERL83620.1 CH940647 EDW70777.1 KRF85103.1 GL766526 EFZ15067.1 CVRI01000036 CRK92954.1 MRZV01000111 PIK58497.1 DS232168 EDS36536.1 NEVH01005303 PNF38722.1 KQ983106 KYQ47415.1
SOQ49649.1 GDQN01003165 GDQN01000896 JAT87889.1 JAT90158.1 GDQN01011586 JAT79468.1 GDQN01004039 JAT87015.1 GDQN01006909 JAT84145.1 JTDY01001022 KOB75106.1 AGBW02009960 OWR49480.1 GAIX01000007 JAA92553.1 KQ459606 KPI91455.1 KQ460398 KPJ15072.1 GALX01001820 JAB66646.1 GALX01001819 JAB66647.1 KQ971371 KYB25520.1 EFA09968.1 AXCN02000934 GEBQ01008172 JAT31805.1 GEBQ01021625 JAT18352.1 GEBQ01000597 JAT39380.1 AXCM01007726 AAAB01008960 EAA10870.4 APCN01004380 KB632204 ERL90108.1 GECZ01003808 JAS65961.1 BT127651 AEE62613.1 APGK01043377 APGK01043378 KB741014 ENN75454.1 GECU01011989 JAS95717.1 GECU01015943 JAS91763.1 GECU01006492 JAT01215.1 KK852687 KDR18392.1 JXUM01062867 KQ562219 KXJ76368.1 GDAI01000038 JAI17565.1 GAHY01001982 JAA75528.1 GBRD01011318 JAG54506.1 GBHO01011531 JAG32073.1 PYGN01000877 PSN39773.1 GECU01010790 JAS96916.1 GGFM01003820 MBW24571.1 GBGD01002295 JAC86594.1 GGFK01006939 MBW40260.1 GGFJ01008382 MBW57523.1 ATLV01022985 KE525339 KFB48211.1 KQ435840 KOX71397.1 GGFM01004269 MBW25020.1 GGFJ01007267 MBW56408.1 GGFJ01007317 MBW56458.1 GL448543 EFN84334.1 CH477404 EAT41655.1 EAT41656.1 GL438827 EFN68263.1 GGFL01000336 MBW64514.1 GFDL01014101 JAV20944.1 GECL01002597 JAP03527.1 GGFL01000335 MBW64513.1 GAMC01011192 JAB95363.1 GAMC01011194 JAB95361.1 GEZM01054604 JAV73530.1 GGFM01003863 MBW24614.1 GFWV01018742 MAA43470.1 GEZM01054603 JAV73531.1 ADMH02001054 ETN64294.1 GFDL01013205 JAV21840.1 AAZX01003332 KZ288204 PBC33222.1 KA646705 AFP61334.1 GEDC01010289 JAS27009.1 GEDC01022114 JAS15184.1 KK854028 PTY09784.1 KB630474 ERL83620.1 CH940647 EDW70777.1 KRF85103.1 GL766526 EFZ15067.1 CVRI01000036 CRK92954.1 MRZV01000111 PIK58497.1 DS232168 EDS36536.1 NEVH01005303 PNF38722.1 KQ983106 KYQ47415.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053268
UP000053240
+ More
UP000007266 UP000075920 UP000075886 UP000075885 UP000075883 UP000007062 UP000075882 UP000075840 UP000076407 UP000030742 UP000075900 UP000075902 UP000075903 UP000019118 UP000075881 UP000027135 UP000069940 UP000249989 UP000245037 UP000076408 UP000075880 UP000030765 UP000053105 UP000008237 UP000075901 UP000069272 UP000008820 UP000000311 UP000075884 UP000000673 UP000002358 UP000242457 UP000095301 UP000008792 UP000183832 UP000230750 UP000002320 UP000005203 UP000235965 UP000095300 UP000075809
UP000007266 UP000075920 UP000075886 UP000075885 UP000075883 UP000007062 UP000075882 UP000075840 UP000076407 UP000030742 UP000075900 UP000075902 UP000075903 UP000019118 UP000075881 UP000027135 UP000069940 UP000249989 UP000245037 UP000076408 UP000075880 UP000030765 UP000053105 UP000008237 UP000075901 UP000069272 UP000008820 UP000000311 UP000075884 UP000000673 UP000002358 UP000242457 UP000095301 UP000008792 UP000183832 UP000230750 UP000002320 UP000005203 UP000235965 UP000095300 UP000075809
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JBH1
A0A2A4JAK5
A0A2A4J9T5
A0A2A4JB18
A0A2H1W975
A0A1E1WLK2
+ More
A0A1E1VXK7 A0A1E1WJ44 A0A1E1WB24 A0A0L7LIS2 A0A212F6X0 S4PPU5 A0A194PK35 A0A194RBU6 V5IA46 V5GXW2 A0A139WC74 A0A182WLT8 D6X1S7 A0A182Q9I0 A0A1B6M7B0 A0A1B6L3P0 A0A1B6MTX9 A0A182PVJ3 A0A182MCG9 Q7Q7A3 A0A182LJT6 A0A182HM92 A0A182XCW3 U4UCQ8 A0A1B6GU67 J3JWN3 A0A182RC29 A0A182TXA8 A0A182VBV3 N6T5H5 A0A182JV20 A0A1B6J935 A0A1B6IXX8 A0A1B6JPP6 A0A067R539 A0A182GH13 A0A0K8TUE9 R4G7X1 A0A0K8SMG8 A0A0A9YLU8 A0A2P8Y696 A0A1B6JCC8 A0A182Y0R2 A0A182J9X7 A0A2M3Z7U1 A0A069DYJ2 A0A2M4AHK6 A0A2M4BWW4 A0A084WDB7 A0A0M8ZUS9 A0A2M3Z958 A0A2M4BUH2 A0A2M4BUM1 E2BIY9 A0A182ST72 A0A182FHP3 Q174X6 A0A1S4FEE5 E2AE30 A0A2M4CGS7 A0A1Q3F070 A0A0V0G6A9 A0A2M4CGS1 W8BEE4 A0A182NMD4 W8BPB6 A0A1Y1LME9 A0A2M3Z7X0 A0A293MVX7 A0A1Y1LKT5 W5JNE3 A0A1Q3F2Q5 K7IT22 A0A2A3EPM9 A0A1I8ND92 T1PEX8 A0A1B6DMX2 A0A1B6CP86 A0A2R7VQI7 A0A1I8ND98 U4U0P6 B4LIJ0 A0A0Q9WV42 E9IWI1 A0A1J1HZK0 A0A2G8LE52 B0WXK6 A0A088ACJ5 A0A2J7RD15 A0A1I8Q993 A0A151WHZ0
A0A1E1VXK7 A0A1E1WJ44 A0A1E1WB24 A0A0L7LIS2 A0A212F6X0 S4PPU5 A0A194PK35 A0A194RBU6 V5IA46 V5GXW2 A0A139WC74 A0A182WLT8 D6X1S7 A0A182Q9I0 A0A1B6M7B0 A0A1B6L3P0 A0A1B6MTX9 A0A182PVJ3 A0A182MCG9 Q7Q7A3 A0A182LJT6 A0A182HM92 A0A182XCW3 U4UCQ8 A0A1B6GU67 J3JWN3 A0A182RC29 A0A182TXA8 A0A182VBV3 N6T5H5 A0A182JV20 A0A1B6J935 A0A1B6IXX8 A0A1B6JPP6 A0A067R539 A0A182GH13 A0A0K8TUE9 R4G7X1 A0A0K8SMG8 A0A0A9YLU8 A0A2P8Y696 A0A1B6JCC8 A0A182Y0R2 A0A182J9X7 A0A2M3Z7U1 A0A069DYJ2 A0A2M4AHK6 A0A2M4BWW4 A0A084WDB7 A0A0M8ZUS9 A0A2M3Z958 A0A2M4BUH2 A0A2M4BUM1 E2BIY9 A0A182ST72 A0A182FHP3 Q174X6 A0A1S4FEE5 E2AE30 A0A2M4CGS7 A0A1Q3F070 A0A0V0G6A9 A0A2M4CGS1 W8BEE4 A0A182NMD4 W8BPB6 A0A1Y1LME9 A0A2M3Z7X0 A0A293MVX7 A0A1Y1LKT5 W5JNE3 A0A1Q3F2Q5 K7IT22 A0A2A3EPM9 A0A1I8ND92 T1PEX8 A0A1B6DMX2 A0A1B6CP86 A0A2R7VQI7 A0A1I8ND98 U4U0P6 B4LIJ0 A0A0Q9WV42 E9IWI1 A0A1J1HZK0 A0A2G8LE52 B0WXK6 A0A088ACJ5 A0A2J7RD15 A0A1I8Q993 A0A151WHZ0
PDB
5Y6L
E-value=0.00013874,
Score=105
Ontologies
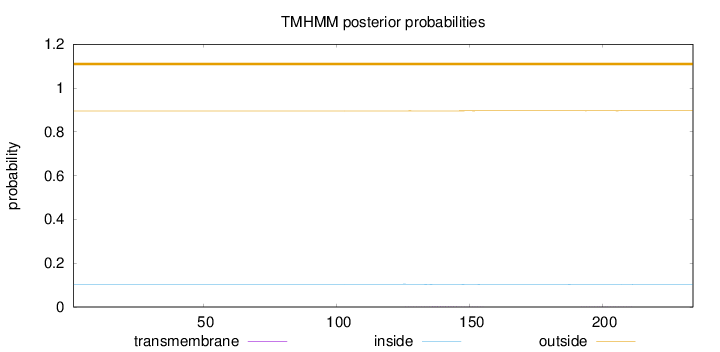
Topology
Length:
234
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04256
Exp number, first 60 AAs:
0.00071
Total prob of N-in:
0.10434
outside
1 - 234
Population Genetic Test Statistics
Pi
170.476147
Theta
174.698269
Tajima's D
0.000465
CLR
1.826933
CSRT
0.368631568421579
Interpretation
Uncertain
