Pre Gene Modal
BGIBMGA006865
Annotation
cAMP_responsive_element_binding_protein_[Bombyx_mori]
Full name
Annexin
Transcription factor
Location in the cell
Nuclear Reliability : 4.588
Sequence
CDS
ATGGAAGGGATGGTTGAAGAGAACGGCACCACAGGCGGTGCTGGCACGACGGATCCACTGGCCAGCGCGAGCTCTTCGACACCGGGTTCCACTCCTCATGTTGTCACCAGTATAGTACAACTTACTCTACCCAGTCAGGCTCCCTCTGCACAGGTGCAGTCTGTTATTCAACCTAATCAACAATCAGTGATACAAACAGCATCCAATATCCAATCTGTACAGATACCAAAGGGAAATGTTATACTTGTTAGTAAACCCAGTTCTGTAATACACACTACGCAAGGGACATTACAAACGTTACAGATAAAACCAGAACCTAATACACTTGTGAATACACAAGGTCAGTCATGTAGTGATGAAAGCTGTGGCAGTGGTGATGAAAGTCCCAAACGTAAATATAGAGAAATGTTGACTAGACGTCCGTCTTATCGAAAAATTTTAAATGACCTTGGAGGAGCAGAAATTGCAGTCATTCCAGCTGGAACTATACAAACTGACAGCGGATTGCACACGTTGGCCGTGTCCGGAGCGGCGGGGGGCGGAGCCATAGTGCAGTATGCAGCGAATCAAGATGGACAATTCTATGTACCGGGTCCCATACTAGAAGACCAAACGCGCAAACGAGAACTACGACTAATGAAAAACCGAGAAGCCGCTAGGGAATGCCGCAGAAAGAAAAAGGAGTACATCAAGTGCCTTGAAAACAGAGTTGCAGTCCTTGAAAACCAAAATAAAGCGCTAATAGAAGAGTTGAAGACATTAAAGGAATTATATTGCCAACAGAAAACGGAATGA
Protein
MEGMVEENGTTGGAGTTDPLASASSSTPGSTPHVVTSIVQLTLPSQAPSAQVQSVIQPNQQSVIQTASNIQSVQIPKGNVILVSKPSSVIHTTQGTLQTLQIKPEPNTLVNTQGQSCSDESCGSGDESPKRKYREMLTRRPSYRKILNDLGGAEIAVIPAGTIQTDSGLHTLAVSGAAGGGAIVQYAANQDGQFYVPGPILEDQTRKRELRLMKNREAARECRRKKKEYIKCLENRVAVLENQNKALIEELKTLKELYCQQKTE
Summary
Similarity
Belongs to the annexin family.
Uniprot
Q2F613
A9XTS8
E9JEH1
A0A0S2VHW9
A0A3G1CVR7
A0A194PDX1
+ More
A0A2H1W9Y2 A0A194RC79 A0A212F6W5 A0A2A4JWH5 A0A3S2TC30 A9XTS9 A0A0L7LRV0 S4PC90 B6DRC3 B6DRD2 B6DRC1 B6DRD0 A0A1W4X6G5 A0A3L8DLX0 A0A026W4V0 Q8MU62 A0A232FIG9 E0W418 B6DRC6 B6DRD5 A0A1W4X6B2 Q8MYD1 E2BTD2 B6DRC0 B6DRC9 A0A1W4WVM8 A0A1W4X525 Q8MU64 A0A1B6J1E0 B6DRB9 B6DRC8 B2CKL8 S4VQR8 U5EXI8 A0A1B6IQQ1 Q8MU63 A0A1B6KHG7 A0A1W4WVM5 Q8MU69 A0A1B6EU58 B0W9J4 A0A182GP16 A0A1B6IW03 A0A1Q3F9H4 A0A1Q3FA83 A0A2A3E584 Q8MU65 A0A1B6I611 K7IT30 A0A154NYU1 Q8MYC9 Q8MU66 A0A088A5B7 A0A1S4G6I6 Q8MTP6 Q8MYC8 A0A1Q3F9H1 A0A1Q3F9Y1 A0A151X0H2 A0A195D6L3 Q8MYD0 Q8MYC7 A0A1W4X6H0 D0E8D3 D0E8D1 Q8MYC5 A0A151J4D6 A0A1W4WVL2 B6DRC2 B6DRD1 E1ZYA7 T1E284 A0A0A9XI42 A0A1L8DIH8 A0A182XY15 A0A146LD13 A0A182PR30 A0A1Q3F9I8 A0A1Q3F9Z1 A0A195BRP8 A0A0A9XFZ4 A0A1Q3F963 A0A1Q3FAA7 A0A067RH93 A0A0K8SPP6 A0A182W5A0 A0A0B8RSM5 Q8MU68 A0A1B6CBU9 A0A0A9XRD8 A0A2J7R725 Q8MYC4 A0A0A9XLH1 A0A336JX45
A0A2H1W9Y2 A0A194RC79 A0A212F6W5 A0A2A4JWH5 A0A3S2TC30 A9XTS9 A0A0L7LRV0 S4PC90 B6DRC3 B6DRD2 B6DRC1 B6DRD0 A0A1W4X6G5 A0A3L8DLX0 A0A026W4V0 Q8MU62 A0A232FIG9 E0W418 B6DRC6 B6DRD5 A0A1W4X6B2 Q8MYD1 E2BTD2 B6DRC0 B6DRC9 A0A1W4WVM8 A0A1W4X525 Q8MU64 A0A1B6J1E0 B6DRB9 B6DRC8 B2CKL8 S4VQR8 U5EXI8 A0A1B6IQQ1 Q8MU63 A0A1B6KHG7 A0A1W4WVM5 Q8MU69 A0A1B6EU58 B0W9J4 A0A182GP16 A0A1B6IW03 A0A1Q3F9H4 A0A1Q3FA83 A0A2A3E584 Q8MU65 A0A1B6I611 K7IT30 A0A154NYU1 Q8MYC9 Q8MU66 A0A088A5B7 A0A1S4G6I6 Q8MTP6 Q8MYC8 A0A1Q3F9H1 A0A1Q3F9Y1 A0A151X0H2 A0A195D6L3 Q8MYD0 Q8MYC7 A0A1W4X6H0 D0E8D3 D0E8D1 Q8MYC5 A0A151J4D6 A0A1W4WVL2 B6DRC2 B6DRD1 E1ZYA7 T1E284 A0A0A9XI42 A0A1L8DIH8 A0A182XY15 A0A146LD13 A0A182PR30 A0A1Q3F9I8 A0A1Q3F9Z1 A0A195BRP8 A0A0A9XFZ4 A0A1Q3F963 A0A1Q3FAA7 A0A067RH93 A0A0K8SPP6 A0A182W5A0 A0A0B8RSM5 Q8MU68 A0A1B6CBU9 A0A0A9XRD8 A0A2J7R725 Q8MYC4 A0A0A9XLH1 A0A336JX45
Pubmed
EMBL
DQ311259
EF682070
ABD36204.1
ABV48888.1
EF682068
ABV48886.1
+ More
BABH01030896 GQ426288 ADM32514.1 KT207931 ALP86606.1 KU552108 AMH87784.1 KQ459606 KPI91457.1 ODYU01007140 SOQ49652.1 KQ460398 KPJ15074.1 AGBW02009960 OWR49482.1 NWSH01000473 PCG76169.1 RSAL01000479 RVE41583.1 EF682069 ABV48887.1 JTDY01000228 KOB78190.1 GAIX01004281 JAA88279.1 FJ155911 ACI03618.1 FJ155920 ACI03627.1 FJ155909 ACI03616.1 FJ155918 ACI03625.1 QOIP01000006 RLU21425.1 KK107485 EZA50079.1 AJ430466 CAD23079.1 NNAY01000179 OXU30259.1 DS235886 EEB20374.1 FJ155914 ACI03621.1 FJ155923 ACI03630.1 AJ430615 AJ430582 CAD24865.1 GL450389 EFN81053.1 FJ155908 ACI03615.1 FJ155917 ACI03624.1 AJ430464 CAD23077.1 GECU01014723 GECU01002736 JAS92983.1 JAT04971.1 FJ155907 ACI03614.1 FJ155916 ACI03623.1 EU523223 ACB20690.1 KC814690 AGO81725.1 GANO01000929 JAB58942.1 GECU01018461 GECU01014536 GECU01007078 JAS89245.1 JAS93170.1 JAT00629.1 AJ430465 CAD23078.1 GEBQ01029108 GEBQ01012279 GEBQ01001983 JAT10869.1 JAT27698.1 JAT37994.1 AJ430459 CAD23072.1 GECZ01028235 JAS41534.1 DS231864 EDS40219.1 JXUM01077749 JXUM01077750 JXUM01077751 KQ563026 KXJ74619.1 GECU01016587 GECU01007086 JAS91119.1 JAT00621.1 GFDL01010876 JAV24169.1 GFDL01010627 JAV24418.1 KZ288364 PBC26855.1 AJ430463 CAD23076.1 GECU01025359 JAS82347.1 AAZX01008089 KQ434783 KZC04793.1 CAD24867.1 AJ430462 CAD23075.1 AY083158 AAL92477.1 CAD24868.1 GFDL01010863 JAV24182.1 GFDL01010737 JAV24308.1 KQ982622 KYQ53733.1 KQ976760 KYN08535.1 CAD24866.1 CAD24869.1 GQ411372 GQ411376 ACX54102.1 GQ411375 ACX54101.1 CAD24871.1 KQ980151 KYN17494.1 FJ155910 ACI03617.1 FJ155919 ACI03626.1 GL435204 EFN73806.1 GALA01000969 JAA93883.1 GBHO01023985 GBHO01023984 JAG19619.1 JAG19620.1 GFDF01007899 JAV06185.1 GDHC01012538 GDHC01003627 JAQ06091.1 JAQ15002.1 GFDL01010815 JAV24230.1 GFDL01010654 JAV24391.1 KQ976417 KYM89690.1 GBHO01023987 JAG19617.1 GFDL01010930 JAV24115.1 GFDL01010539 JAV24506.1 KK852470 KDR23211.1 GBRD01010576 JAG55248.1 GBRD01005629 JAG60192.1 AJ430460 CAD23073.1 GEDC01031040 GEDC01026360 JAS06258.1 JAS10938.1 GBHO01023990 GBHO01023989 GBRD01005630 JAG19614.1 JAG19615.1 JAG60191.1 NEVH01006738 PNF36633.1 CAD24872.1 GBHO01023986 GBRD01005628 JAG19618.1 JAG60193.1 UFQS01000015 UFQT01000015 SSW97356.1 SSX17742.1
BABH01030896 GQ426288 ADM32514.1 KT207931 ALP86606.1 KU552108 AMH87784.1 KQ459606 KPI91457.1 ODYU01007140 SOQ49652.1 KQ460398 KPJ15074.1 AGBW02009960 OWR49482.1 NWSH01000473 PCG76169.1 RSAL01000479 RVE41583.1 EF682069 ABV48887.1 JTDY01000228 KOB78190.1 GAIX01004281 JAA88279.1 FJ155911 ACI03618.1 FJ155920 ACI03627.1 FJ155909 ACI03616.1 FJ155918 ACI03625.1 QOIP01000006 RLU21425.1 KK107485 EZA50079.1 AJ430466 CAD23079.1 NNAY01000179 OXU30259.1 DS235886 EEB20374.1 FJ155914 ACI03621.1 FJ155923 ACI03630.1 AJ430615 AJ430582 CAD24865.1 GL450389 EFN81053.1 FJ155908 ACI03615.1 FJ155917 ACI03624.1 AJ430464 CAD23077.1 GECU01014723 GECU01002736 JAS92983.1 JAT04971.1 FJ155907 ACI03614.1 FJ155916 ACI03623.1 EU523223 ACB20690.1 KC814690 AGO81725.1 GANO01000929 JAB58942.1 GECU01018461 GECU01014536 GECU01007078 JAS89245.1 JAS93170.1 JAT00629.1 AJ430465 CAD23078.1 GEBQ01029108 GEBQ01012279 GEBQ01001983 JAT10869.1 JAT27698.1 JAT37994.1 AJ430459 CAD23072.1 GECZ01028235 JAS41534.1 DS231864 EDS40219.1 JXUM01077749 JXUM01077750 JXUM01077751 KQ563026 KXJ74619.1 GECU01016587 GECU01007086 JAS91119.1 JAT00621.1 GFDL01010876 JAV24169.1 GFDL01010627 JAV24418.1 KZ288364 PBC26855.1 AJ430463 CAD23076.1 GECU01025359 JAS82347.1 AAZX01008089 KQ434783 KZC04793.1 CAD24867.1 AJ430462 CAD23075.1 AY083158 AAL92477.1 CAD24868.1 GFDL01010863 JAV24182.1 GFDL01010737 JAV24308.1 KQ982622 KYQ53733.1 KQ976760 KYN08535.1 CAD24866.1 CAD24869.1 GQ411372 GQ411376 ACX54102.1 GQ411375 ACX54101.1 CAD24871.1 KQ980151 KYN17494.1 FJ155910 ACI03617.1 FJ155919 ACI03626.1 GL435204 EFN73806.1 GALA01000969 JAA93883.1 GBHO01023985 GBHO01023984 JAG19619.1 JAG19620.1 GFDF01007899 JAV06185.1 GDHC01012538 GDHC01003627 JAQ06091.1 JAQ15002.1 GFDL01010815 JAV24230.1 GFDL01010654 JAV24391.1 KQ976417 KYM89690.1 GBHO01023987 JAG19617.1 GFDL01010930 JAV24115.1 GFDL01010539 JAV24506.1 KK852470 KDR23211.1 GBRD01010576 JAG55248.1 GBRD01005629 JAG60192.1 AJ430460 CAD23073.1 GEDC01031040 GEDC01026360 JAS06258.1 JAS10938.1 GBHO01023990 GBHO01023989 GBRD01005630 JAG19614.1 JAG19615.1 JAG60191.1 NEVH01006738 PNF36633.1 CAD24872.1 GBHO01023986 GBRD01005628 JAG19618.1 JAG60193.1 UFQS01000015 UFQT01000015 SSW97356.1 SSX17742.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000283053
+ More
UP000037510 UP000192223 UP000279307 UP000053097 UP000215335 UP000009046 UP000008237 UP000002320 UP000069940 UP000249989 UP000242457 UP000002358 UP000076502 UP000005203 UP000075809 UP000078542 UP000078492 UP000000311 UP000076408 UP000075885 UP000078540 UP000027135 UP000075920 UP000235965
UP000037510 UP000192223 UP000279307 UP000053097 UP000215335 UP000009046 UP000008237 UP000002320 UP000069940 UP000249989 UP000242457 UP000002358 UP000076502 UP000005203 UP000075809 UP000078542 UP000078492 UP000000311 UP000076408 UP000075885 UP000078540 UP000027135 UP000075920 UP000235965
Interpro
Gene 3D
ProteinModelPortal
Q2F613
A9XTS8
E9JEH1
A0A0S2VHW9
A0A3G1CVR7
A0A194PDX1
+ More
A0A2H1W9Y2 A0A194RC79 A0A212F6W5 A0A2A4JWH5 A0A3S2TC30 A9XTS9 A0A0L7LRV0 S4PC90 B6DRC3 B6DRD2 B6DRC1 B6DRD0 A0A1W4X6G5 A0A3L8DLX0 A0A026W4V0 Q8MU62 A0A232FIG9 E0W418 B6DRC6 B6DRD5 A0A1W4X6B2 Q8MYD1 E2BTD2 B6DRC0 B6DRC9 A0A1W4WVM8 A0A1W4X525 Q8MU64 A0A1B6J1E0 B6DRB9 B6DRC8 B2CKL8 S4VQR8 U5EXI8 A0A1B6IQQ1 Q8MU63 A0A1B6KHG7 A0A1W4WVM5 Q8MU69 A0A1B6EU58 B0W9J4 A0A182GP16 A0A1B6IW03 A0A1Q3F9H4 A0A1Q3FA83 A0A2A3E584 Q8MU65 A0A1B6I611 K7IT30 A0A154NYU1 Q8MYC9 Q8MU66 A0A088A5B7 A0A1S4G6I6 Q8MTP6 Q8MYC8 A0A1Q3F9H1 A0A1Q3F9Y1 A0A151X0H2 A0A195D6L3 Q8MYD0 Q8MYC7 A0A1W4X6H0 D0E8D3 D0E8D1 Q8MYC5 A0A151J4D6 A0A1W4WVL2 B6DRC2 B6DRD1 E1ZYA7 T1E284 A0A0A9XI42 A0A1L8DIH8 A0A182XY15 A0A146LD13 A0A182PR30 A0A1Q3F9I8 A0A1Q3F9Z1 A0A195BRP8 A0A0A9XFZ4 A0A1Q3F963 A0A1Q3FAA7 A0A067RH93 A0A0K8SPP6 A0A182W5A0 A0A0B8RSM5 Q8MU68 A0A1B6CBU9 A0A0A9XRD8 A0A2J7R725 Q8MYC4 A0A0A9XLH1 A0A336JX45
A0A2H1W9Y2 A0A194RC79 A0A212F6W5 A0A2A4JWH5 A0A3S2TC30 A9XTS9 A0A0L7LRV0 S4PC90 B6DRC3 B6DRD2 B6DRC1 B6DRD0 A0A1W4X6G5 A0A3L8DLX0 A0A026W4V0 Q8MU62 A0A232FIG9 E0W418 B6DRC6 B6DRD5 A0A1W4X6B2 Q8MYD1 E2BTD2 B6DRC0 B6DRC9 A0A1W4WVM8 A0A1W4X525 Q8MU64 A0A1B6J1E0 B6DRB9 B6DRC8 B2CKL8 S4VQR8 U5EXI8 A0A1B6IQQ1 Q8MU63 A0A1B6KHG7 A0A1W4WVM5 Q8MU69 A0A1B6EU58 B0W9J4 A0A182GP16 A0A1B6IW03 A0A1Q3F9H4 A0A1Q3FA83 A0A2A3E584 Q8MU65 A0A1B6I611 K7IT30 A0A154NYU1 Q8MYC9 Q8MU66 A0A088A5B7 A0A1S4G6I6 Q8MTP6 Q8MYC8 A0A1Q3F9H1 A0A1Q3F9Y1 A0A151X0H2 A0A195D6L3 Q8MYD0 Q8MYC7 A0A1W4X6H0 D0E8D3 D0E8D1 Q8MYC5 A0A151J4D6 A0A1W4WVL2 B6DRC2 B6DRD1 E1ZYA7 T1E284 A0A0A9XI42 A0A1L8DIH8 A0A182XY15 A0A146LD13 A0A182PR30 A0A1Q3F9I8 A0A1Q3F9Z1 A0A195BRP8 A0A0A9XFZ4 A0A1Q3F963 A0A1Q3FAA7 A0A067RH93 A0A0K8SPP6 A0A182W5A0 A0A0B8RSM5 Q8MU68 A0A1B6CBU9 A0A0A9XRD8 A0A2J7R725 Q8MYC4 A0A0A9XLH1 A0A336JX45
PDB
5ZKO
E-value=9.0081e-12,
Score=167
Ontologies
GO
Topology
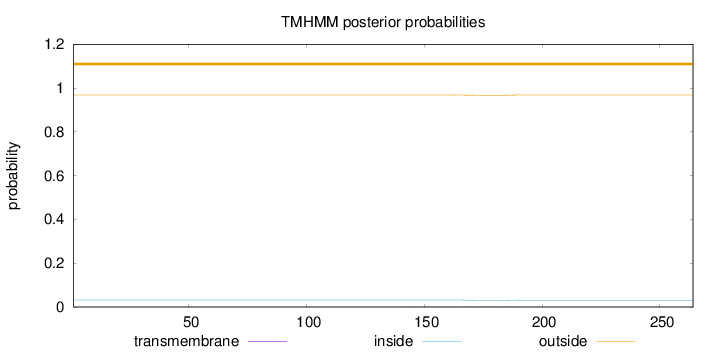
Length:
264
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06098
Exp number, first 60 AAs:
0.00043
Total prob of N-in:
0.03223
outside
1 - 264
Population Genetic Test Statistics
Pi
259.377293
Theta
209.897232
Tajima's D
0.776674
CLR
0.023301
CSRT
0.591720413979301
Interpretation
Uncertain
