Gene
KWMTBOMO05713 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006823
Annotation
4-aminobutyrate_aminotransferase_[Chilo_suppressalis]
Location in the cell
Cytoplasmic Reliability : 2.147 Mitochondrial Reliability : 1.588
Sequence
CDS
ATGTTGTTACGAACTTCAAGAAGTGCTCACGTTTTTAGAGGTGTTAGGGCTTATTCGGTTTTGCAGCATGAACCAGATAAACCCAATATTCAAACAGCGATTCCCGGGCCCAAGTCGCAGCAGCTGCTGAAAGAACTCAACACTCTACAGCAAGCGGGAGCAGTTCAGCTTTTTGCTGATTATGACAAAAGTATTGGAAATTATTTCTTTGACGCCGATGGTAATGCATTCCTAGATGCTTTCACACAAATATCTTCACTTCCCGTGGGGTATAATCATCCTGAATTATTAAGCGCATTTGAAGATCAACACAATCTTAAATCTTTGATTAATAGGCCAGCTCTGGGAGTTTTTCCTTCAGCTGATTGGCCAGAGAAATTGAAAAATGTGTTATTATCTGTTGGACCCGTTGGGTTGGACAACATAGCGACAATGATGTGTGGCTCCTGTTCTATTGAGAATGCGTATAAAACTGTATTCATATGGTATCAGACCCGGGAACGCGGAGGAAAACTGGATTTCACACCCGAAGAAATTGACAGTTGTATGTTGAATCAAGTTCCTGGATCACCTAAACTGTCAATATTATCATTTGAGGGAAGTTTCCACGGTCGAACATTTGGAGCTCTGTCTACTACTAGATCGAAGCCATTGCATAAGTTAGACTGTCCAGCTTTTGATTGGCCGGTTGCTCCGTTTCCCAGATACAAATATCCCCTTGAAGAAAATATTAATGAAAATAAGCAAGAAGACAAGAGATGTCTTGAGCAAGTGGCTGACCTAATTGAGAAGTATAAAAAAAAGGGCAATCCGGTTGCTGGTATTGTTGTGGAACCAATTCAGTCCGAAGGAGGTGATAATGAAGCTTCACCGGAATTCTTTCGAGACCTTCAAAAATTATCTAAGGAGAAGGCGGTTGCATTTATTGTTGATGAAGTGCAAACCGGATGTGGTCCAACCGGGAAAATGTGGTGTCATGAACACTTCGATCTACCGACGTCACCCGACGTCGTCACCTTTAGCAAGAAAATGTTGACTGGAGGATTCTACTTCACTGCTGATTTTAAACCTCCGCACGCGTACAGAGTTTTCAACACGTGGATGGGGGATCCAGGTAAATTGATTTTACTCGAGCGCGTTCTCAAGGTCATCAAACAGGAAAACTTGCTGGATTTAGTCAACAAGACCGGAAACGTTTTGAAAAATGGCCTACACGATCTGGAAAAGGAGTTCCCTGGAGTAATACATAGTGTTCGTGGGCGAGGCACGTTCTTGGCTTATAATGCTCCCACTACTGAAACAAGGGATAAGATAAACGCCGGATTAAAGAAAAACGGTGTTTTGGGGGGAGTATGTGGTGTACGTGCAATTCGTCTGAGACCAGCATTAATTTTCGAACCTAAACATGCTGCTATTTATTTGGATGTTCTGCGGAAGACCTTGAAAGAAATATAA
Protein
MLLRTSRSAHVFRGVRAYSVLQHEPDKPNIQTAIPGPKSQQLLKELNTLQQAGAVQLFADYDKSIGNYFFDADGNAFLDAFTQISSLPVGYNHPELLSAFEDQHNLKSLINRPALGVFPSADWPEKLKNVLLSVGPVGLDNIATMMCGSCSIENAYKTVFIWYQTRERGGKLDFTPEEIDSCMLNQVPGSPKLSILSFEGSFHGRTFGALSTTRSKPLHKLDCPAFDWPVAPFPRYKYPLEENINENKQEDKRCLEQVADLIEKYKKKGNPVAGIVVEPIQSEGGDNEASPEFFRDLQKLSKEKAVAFIVDEVQTGCGPTGKMWCHEHFDLPTSPDVVTFSKKMLTGGFYFTADFKPPHAYRVFNTWMGDPGKLILLERVLKVIKQENLLDLVNKTGNVLKNGLHDLEKEFPGVIHSVRGRGTFLAYNAPTTETRDKINAGLKKNGVLGGVCGVRAIRLRPALIFEPKHAAIYLDVLRKTLKEI
Summary
Similarity
Belongs to the class-III pyridoxal-phosphate-dependent aminotransferase family.
Uniprot
H9JBC8
A0A0G3VJL6
A0A212F6Y5
A0A194RC84
A0A194PDX6
A0A240PK91
+ More
A0A067QVN2 A0A1Q3G538 A0A182J9A8 A0A2A4JXQ1 Q16LL9 A0A1L8E2E4 W5JFY4 A0A182Y2E1 A0A1I8PPP6 A0A1Y1MBC3 A0A084WF13 A0A1I8MVN3 A0A1B6FKA1 A0A336M3B2 A0A2S2NV32 A0A1Y9HDW1 A0A182QNT6 A0A1Y9IRY4 A0A1Y9J0U0 A0A2C9GTZ2 Q7QIN5 A0A1Y9IW69 J9JYS0 A0A0L0BVX1 A0A1Y9GKT6 A0A2C9GH02 A0A1Y9H2G8 B4HBS9 B4KXQ2 A0A0A9YM18 J3JUS5 A0A0Q9XQQ8 A0A2H1VLC9 A0A1D2NLF3 A0A1J1HZN1 A0A0M4F289 B4LFN4 W8B5U3 A0A069DUY6 A0A139WC59 A0A182MRV7 A0A139WCG1 A0A023F4I3 A0A1W4WNP7 A0A224XJH7 A0A182SCB3 F4WEK9 B4MXN9 B4J1R6 A0A2H8TS20 A0A195FNA0 A0A1B6EGL6 E2AE16 A0A0A1XD99 A0A0M9A1F2 A0A0L7R1B0 A0A151WHT6 A0A195E897 A0A1W4W7M4 B4QRE7 B4PFU5 A0A026WA63 A0A034VEP3 Q9VW68 A0A3L8DE55 T1GPX3 A0A158NF28 A0A0K8VJY6 B3M8R4 B4IIU2 B3NE22 Q8MYV0 E0VMX4 A0A088AJD7 A0A2A3E4C1 A0A1B6GQH7 A0A310SS18 E2AE15 A0A1B6JAS3 K7IT15 A0A3B0KG48 A0A1B6J5S5 A0A1A9XXW5 A0A1A9VCA2 A0A1B0BZA6 A0A232F5Y7 A0A0P4VZ80 E2C9Y8 A0A1B0CN94 A0A1B0GFC3 A0A1B0A395 T1HTC7 A0A1A9X2H8 A0A1A9XPT9
A0A067QVN2 A0A1Q3G538 A0A182J9A8 A0A2A4JXQ1 Q16LL9 A0A1L8E2E4 W5JFY4 A0A182Y2E1 A0A1I8PPP6 A0A1Y1MBC3 A0A084WF13 A0A1I8MVN3 A0A1B6FKA1 A0A336M3B2 A0A2S2NV32 A0A1Y9HDW1 A0A182QNT6 A0A1Y9IRY4 A0A1Y9J0U0 A0A2C9GTZ2 Q7QIN5 A0A1Y9IW69 J9JYS0 A0A0L0BVX1 A0A1Y9GKT6 A0A2C9GH02 A0A1Y9H2G8 B4HBS9 B4KXQ2 A0A0A9YM18 J3JUS5 A0A0Q9XQQ8 A0A2H1VLC9 A0A1D2NLF3 A0A1J1HZN1 A0A0M4F289 B4LFN4 W8B5U3 A0A069DUY6 A0A139WC59 A0A182MRV7 A0A139WCG1 A0A023F4I3 A0A1W4WNP7 A0A224XJH7 A0A182SCB3 F4WEK9 B4MXN9 B4J1R6 A0A2H8TS20 A0A195FNA0 A0A1B6EGL6 E2AE16 A0A0A1XD99 A0A0M9A1F2 A0A0L7R1B0 A0A151WHT6 A0A195E897 A0A1W4W7M4 B4QRE7 B4PFU5 A0A026WA63 A0A034VEP3 Q9VW68 A0A3L8DE55 T1GPX3 A0A158NF28 A0A0K8VJY6 B3M8R4 B4IIU2 B3NE22 Q8MYV0 E0VMX4 A0A088AJD7 A0A2A3E4C1 A0A1B6GQH7 A0A310SS18 E2AE15 A0A1B6JAS3 K7IT15 A0A3B0KG48 A0A1B6J5S5 A0A1A9XXW5 A0A1A9VCA2 A0A1B0BZA6 A0A232F5Y7 A0A0P4VZ80 E2C9Y8 A0A1B0CN94 A0A1B0GFC3 A0A1B0A395 T1HTC7 A0A1A9X2H8 A0A1A9XPT9
Pubmed
19121390
22118469
26354079
24845553
17510324
20920257
+ More
23761445 25244985 28004739 24438588 25315136 20966253 12364791 14747013 17210077 26108605 17994087 25401762 26823975 22516182 27289101 24495485 26334808 18362917 19820115 25474469 21719571 20798317 25830018 22936249 17550304 24508170 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 21347285 20566863 20075255 28648823 27129103
23761445 25244985 28004739 24438588 25315136 20966253 12364791 14747013 17210077 26108605 17994087 25401762 26823975 22516182 27289101 24495485 26334808 18362917 19820115 25474469 21719571 20798317 25830018 22936249 17550304 24508170 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 21347285 20566863 20075255 28648823 27129103
EMBL
BABH01030880
BABH01030881
BABH01030882
KP657642
RSAL01000229
AKL78867.1
+ More
RVE43905.1 AGBW02009960 OWR49487.1 KQ460398 KPJ15079.1 KQ459606 KPI91462.1 KK853196 KDR09977.1 GFDL01000130 JAV34915.1 NWSH01000473 PCG76162.1 CH477901 EAT35209.1 GFDF01001240 JAV12844.1 ADMH02001626 ETN61729.1 GEZM01037897 JAV81860.1 ATLV01023269 KE525341 KFB48807.1 GECZ01019158 JAS50611.1 UFQT01000292 SSX22887.1 GGMR01008420 MBY21039.1 AXCN02000557 AAAB01008807 EAA04779.4 EDO64744.1 ABLF02015715 JRES01001254 KNC24187.1 APCN01003698 CH479267 EDW39523.1 CH933809 EDW19759.2 GBHO01014019 GBHO01011481 GBRD01004188 GDHC01004507 JAG29585.1 JAG32123.1 JAG61633.1 JAQ14122.1 BT126990 AEE61952.1 KRG06807.1 ODYU01003187 SOQ41606.1 LJIJ01000010 ODN06100.1 CVRI01000036 CRK92986.1 CP012525 ALC44300.1 ALC45195.1 CH940647 EDW70352.2 GAMC01010065 JAB96490.1 GBGD01001377 JAC87512.1 KQ971371 KYB25522.1 AXCM01003051 KYB25521.1 GBBI01002615 JAC16097.1 GFTR01006448 JAW09978.1 GL888103 EGI67372.1 CH963876 EDW76808.2 CH916366 EDV97996.1 GFXV01004587 MBW16392.1 KQ981490 KYN41369.1 GEDC01000268 JAS37030.1 GL438827 EFN68249.1 GBXI01005350 JAD08942.1 KQ435763 KOX75380.1 KQ414668 KOC64629.1 KQ983106 KYQ47401.1 KQ979479 KYN21341.1 CM000363 CM002912 EDX11160.1 KMZ00672.1 CM000159 EDW95242.1 KK107295 EZA52982.1 GAKP01018375 JAC40577.1 AE014296 AAF49081.1 AAN11644.1 ACZ94750.1 AHN58150.1 QOIP01000009 RLU18757.1 CAQQ02011693 CAQQ02011694 CAQQ02011695 ADTU01013722 GDHF01013120 JAI39194.1 CH902618 EDV41065.1 CH480844 EDW49863.1 CH954178 EDV52446.1 AY113591 AAM29596.1 DS235327 EEB14730.1 KZ288409 PBC26106.1 GECZ01005067 JAS64702.1 KQ760688 OAD59355.1 EFN68248.1 GECU01011434 JAS96272.1 AAZX01003289 OUUW01000012 SPP87360.1 GECU01013170 JAS94536.1 JXJN01023043 JXJN01023044 NNAY01000889 OXU26055.1 GDKW01001752 JAI54843.1 GL453916 EFN75225.1 AJWK01019906 AJWK01019907 CCAG010021507 ACPB03015605
RVE43905.1 AGBW02009960 OWR49487.1 KQ460398 KPJ15079.1 KQ459606 KPI91462.1 KK853196 KDR09977.1 GFDL01000130 JAV34915.1 NWSH01000473 PCG76162.1 CH477901 EAT35209.1 GFDF01001240 JAV12844.1 ADMH02001626 ETN61729.1 GEZM01037897 JAV81860.1 ATLV01023269 KE525341 KFB48807.1 GECZ01019158 JAS50611.1 UFQT01000292 SSX22887.1 GGMR01008420 MBY21039.1 AXCN02000557 AAAB01008807 EAA04779.4 EDO64744.1 ABLF02015715 JRES01001254 KNC24187.1 APCN01003698 CH479267 EDW39523.1 CH933809 EDW19759.2 GBHO01014019 GBHO01011481 GBRD01004188 GDHC01004507 JAG29585.1 JAG32123.1 JAG61633.1 JAQ14122.1 BT126990 AEE61952.1 KRG06807.1 ODYU01003187 SOQ41606.1 LJIJ01000010 ODN06100.1 CVRI01000036 CRK92986.1 CP012525 ALC44300.1 ALC45195.1 CH940647 EDW70352.2 GAMC01010065 JAB96490.1 GBGD01001377 JAC87512.1 KQ971371 KYB25522.1 AXCM01003051 KYB25521.1 GBBI01002615 JAC16097.1 GFTR01006448 JAW09978.1 GL888103 EGI67372.1 CH963876 EDW76808.2 CH916366 EDV97996.1 GFXV01004587 MBW16392.1 KQ981490 KYN41369.1 GEDC01000268 JAS37030.1 GL438827 EFN68249.1 GBXI01005350 JAD08942.1 KQ435763 KOX75380.1 KQ414668 KOC64629.1 KQ983106 KYQ47401.1 KQ979479 KYN21341.1 CM000363 CM002912 EDX11160.1 KMZ00672.1 CM000159 EDW95242.1 KK107295 EZA52982.1 GAKP01018375 JAC40577.1 AE014296 AAF49081.1 AAN11644.1 ACZ94750.1 AHN58150.1 QOIP01000009 RLU18757.1 CAQQ02011693 CAQQ02011694 CAQQ02011695 ADTU01013722 GDHF01013120 JAI39194.1 CH902618 EDV41065.1 CH480844 EDW49863.1 CH954178 EDV52446.1 AY113591 AAM29596.1 DS235327 EEB14730.1 KZ288409 PBC26106.1 GECZ01005067 JAS64702.1 KQ760688 OAD59355.1 EFN68248.1 GECU01011434 JAS96272.1 AAZX01003289 OUUW01000012 SPP87360.1 GECU01013170 JAS94536.1 JXJN01023043 JXJN01023044 NNAY01000889 OXU26055.1 GDKW01001752 JAI54843.1 GL453916 EFN75225.1 AJWK01019906 AJWK01019907 CCAG010021507 ACPB03015605
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000075885
+ More
UP000027135 UP000075880 UP000218220 UP000008820 UP000000673 UP000076408 UP000095300 UP000030765 UP000095301 UP000075900 UP000075886 UP000075903 UP000076407 UP000075882 UP000007062 UP000075920 UP000007819 UP000037069 UP000075840 UP000069272 UP000075884 UP000008744 UP000009192 UP000094527 UP000183832 UP000092553 UP000008792 UP000007266 UP000075883 UP000192223 UP000075901 UP000007755 UP000007798 UP000001070 UP000078541 UP000000311 UP000053105 UP000053825 UP000075809 UP000078492 UP000192221 UP000000304 UP000002282 UP000053097 UP000000803 UP000279307 UP000015102 UP000005205 UP000007801 UP000001292 UP000008711 UP000009046 UP000005203 UP000242457 UP000002358 UP000268350 UP000092443 UP000078200 UP000092460 UP000215335 UP000008237 UP000092461 UP000092444 UP000092445 UP000015103 UP000091820
UP000027135 UP000075880 UP000218220 UP000008820 UP000000673 UP000076408 UP000095300 UP000030765 UP000095301 UP000075900 UP000075886 UP000075903 UP000076407 UP000075882 UP000007062 UP000075920 UP000007819 UP000037069 UP000075840 UP000069272 UP000075884 UP000008744 UP000009192 UP000094527 UP000183832 UP000092553 UP000008792 UP000007266 UP000075883 UP000192223 UP000075901 UP000007755 UP000007798 UP000001070 UP000078541 UP000000311 UP000053105 UP000053825 UP000075809 UP000078492 UP000192221 UP000000304 UP000002282 UP000053097 UP000000803 UP000279307 UP000015102 UP000005205 UP000007801 UP000001292 UP000008711 UP000009046 UP000005203 UP000242457 UP000002358 UP000268350 UP000092443 UP000078200 UP000092460 UP000215335 UP000008237 UP000092461 UP000092444 UP000092445 UP000015103 UP000091820
Interpro
IPR015422
PyrdxlP-dep_Trfase_dom1
+ More
IPR015421 PyrdxlP-dep_Trfase_major
IPR015424 PyrdxlP-dep_Trfase
IPR004631 4NH2But_aminotransferase_euk
IPR005814 Aminotrans_3
IPR024420 TRAPP_III_complex_Trs85
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR024678 Kinase_OSR1/WNK_CCT
IPR015421 PyrdxlP-dep_Trfase_major
IPR015424 PyrdxlP-dep_Trfase
IPR004631 4NH2But_aminotransferase_euk
IPR005814 Aminotrans_3
IPR024420 TRAPP_III_complex_Trs85
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR024678 Kinase_OSR1/WNK_CCT
Gene 3D
CDD
ProteinModelPortal
H9JBC8
A0A0G3VJL6
A0A212F6Y5
A0A194RC84
A0A194PDX6
A0A240PK91
+ More
A0A067QVN2 A0A1Q3G538 A0A182J9A8 A0A2A4JXQ1 Q16LL9 A0A1L8E2E4 W5JFY4 A0A182Y2E1 A0A1I8PPP6 A0A1Y1MBC3 A0A084WF13 A0A1I8MVN3 A0A1B6FKA1 A0A336M3B2 A0A2S2NV32 A0A1Y9HDW1 A0A182QNT6 A0A1Y9IRY4 A0A1Y9J0U0 A0A2C9GTZ2 Q7QIN5 A0A1Y9IW69 J9JYS0 A0A0L0BVX1 A0A1Y9GKT6 A0A2C9GH02 A0A1Y9H2G8 B4HBS9 B4KXQ2 A0A0A9YM18 J3JUS5 A0A0Q9XQQ8 A0A2H1VLC9 A0A1D2NLF3 A0A1J1HZN1 A0A0M4F289 B4LFN4 W8B5U3 A0A069DUY6 A0A139WC59 A0A182MRV7 A0A139WCG1 A0A023F4I3 A0A1W4WNP7 A0A224XJH7 A0A182SCB3 F4WEK9 B4MXN9 B4J1R6 A0A2H8TS20 A0A195FNA0 A0A1B6EGL6 E2AE16 A0A0A1XD99 A0A0M9A1F2 A0A0L7R1B0 A0A151WHT6 A0A195E897 A0A1W4W7M4 B4QRE7 B4PFU5 A0A026WA63 A0A034VEP3 Q9VW68 A0A3L8DE55 T1GPX3 A0A158NF28 A0A0K8VJY6 B3M8R4 B4IIU2 B3NE22 Q8MYV0 E0VMX4 A0A088AJD7 A0A2A3E4C1 A0A1B6GQH7 A0A310SS18 E2AE15 A0A1B6JAS3 K7IT15 A0A3B0KG48 A0A1B6J5S5 A0A1A9XXW5 A0A1A9VCA2 A0A1B0BZA6 A0A232F5Y7 A0A0P4VZ80 E2C9Y8 A0A1B0CN94 A0A1B0GFC3 A0A1B0A395 T1HTC7 A0A1A9X2H8 A0A1A9XPT9
A0A067QVN2 A0A1Q3G538 A0A182J9A8 A0A2A4JXQ1 Q16LL9 A0A1L8E2E4 W5JFY4 A0A182Y2E1 A0A1I8PPP6 A0A1Y1MBC3 A0A084WF13 A0A1I8MVN3 A0A1B6FKA1 A0A336M3B2 A0A2S2NV32 A0A1Y9HDW1 A0A182QNT6 A0A1Y9IRY4 A0A1Y9J0U0 A0A2C9GTZ2 Q7QIN5 A0A1Y9IW69 J9JYS0 A0A0L0BVX1 A0A1Y9GKT6 A0A2C9GH02 A0A1Y9H2G8 B4HBS9 B4KXQ2 A0A0A9YM18 J3JUS5 A0A0Q9XQQ8 A0A2H1VLC9 A0A1D2NLF3 A0A1J1HZN1 A0A0M4F289 B4LFN4 W8B5U3 A0A069DUY6 A0A139WC59 A0A182MRV7 A0A139WCG1 A0A023F4I3 A0A1W4WNP7 A0A224XJH7 A0A182SCB3 F4WEK9 B4MXN9 B4J1R6 A0A2H8TS20 A0A195FNA0 A0A1B6EGL6 E2AE16 A0A0A1XD99 A0A0M9A1F2 A0A0L7R1B0 A0A151WHT6 A0A195E897 A0A1W4W7M4 B4QRE7 B4PFU5 A0A026WA63 A0A034VEP3 Q9VW68 A0A3L8DE55 T1GPX3 A0A158NF28 A0A0K8VJY6 B3M8R4 B4IIU2 B3NE22 Q8MYV0 E0VMX4 A0A088AJD7 A0A2A3E4C1 A0A1B6GQH7 A0A310SS18 E2AE15 A0A1B6JAS3 K7IT15 A0A3B0KG48 A0A1B6J5S5 A0A1A9XXW5 A0A1A9VCA2 A0A1B0BZA6 A0A232F5Y7 A0A0P4VZ80 E2C9Y8 A0A1B0CN94 A0A1B0GFC3 A0A1B0A395 T1HTC7 A0A1A9X2H8 A0A1A9XPT9
PDB
1OHY
E-value=1.16352e-152,
Score=1385
Ontologies
PATHWAY
00250
Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
PANTHER
Topology
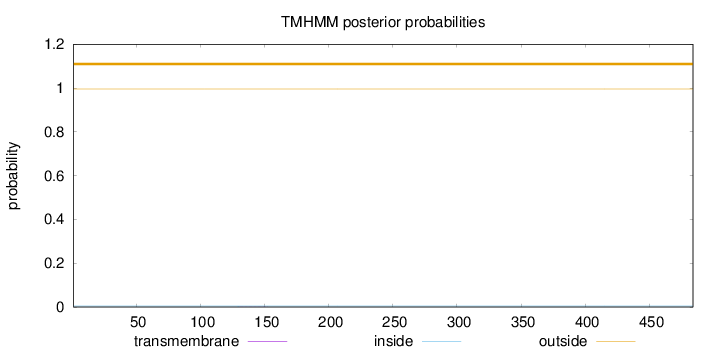
Length:
484
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00633999999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00433
outside
1 - 484
Population Genetic Test Statistics
Pi
23.203393
Theta
180.878765
Tajima's D
1.312263
CLR
0.592532
CSRT
0.741112944352782
Interpretation
Uncertain
