Gene
KWMTBOMO05709
Pre Gene Modal
BGIBMGA006861
Annotation
PREDICTED:_hyccin_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 1.913 PlasmaMembrane Reliability : 1.386
Sequence
CDS
ATGTCTGAGTGGAAGCATCTAATAACTGAATGGCTGAATGAATACACATCTTTGCAAGAAAATGAAGTTCAAAGTTTTGCAGCAGAACATGAACATAATCATGAAATTGCTACAGCCATCTTTAATTTATTTTACTGTGAAGATGAAACTGTAACATATAATTGTGAACTCGAAAATGACGCTCAGCTTGTACAGATATTAGAGAATGTTTGTACACAACTATTTAGTTTCTATAGATCAAAAGAAATACAACTTCAAAGATTTACGTTACAATTCATTCCCACTCTTATATATAATTATTTGAGCTCTGTGGCACAAGGTAAAAAAAAAACATATAGGTGCATAGAAACATTGCTAATTGGCATATACAACTTTGAAGTTGTAGATGAAAATGGAAAACCGAAGGTTGTGTCATTCAGACTTCCATCTTTAGCTCAATCATCAATATATCATGAGCCTCTTTCTTTAGGGGCTCAGTTTTTGACTGAAAGTGCACTGCGTAGATGGGAAGAATGTAATACGAAATTGGTAAATTGGGGCCCGTTGCCACAAGTCGATGTCATCAATGCTCAGAATAGATTGAAAATTGTATCAGCACTTTTATTTGTATACAATCGCCAACTGAGTCTTCTACCTAAACTTTCTTTACGACATTTATGCATTGCAGCTTCAAGAATTGTGACCCATGGTTTTGAAAAAAAGCAAGGAAATGATTCAAAACGTGTTCCAAGAATAGCAGTATCTCCTAGTTTCTTACTAGAGTTGATTGCAGGTGCATACTTTGCTATGTTTAATGAATTCTATACTCTTGCCTTGCAAGCAGTGAAAGATGTCGATCAGAGAGCTCAATATGAATTATTCCCTGAGGTGATGATTGTAACTAGTGCTGTGATAAATTCACTGAAGAATAATGCTTCTGGACAACCATTTGATGGGCCAATGGGTATTAGTGTTGCATTGTCACCAGCAACTACAACGGTTACCATGTCCAAATCTATGATAACAAATGCATCATTCCGCACTAAGAAACTGCCAGACGATATTCCAATACAAGCCGGACAAGCGACAACAGCCGAGTCCACCGATGTGCTGACTTCAATTACCGAGGAAGGCGAATCGGAAACTGCTCCGATACAACGCGGCATAATTGCTCGAAATTCGAAAGCCAAACTATCTTCTTTCCCAGTATTAGGCAAAAAAACAAAGGATACTAAAGATAAGGGTAACGAAAAGAAAGTGATACTGAAGGAATCCTCAAAAGGCATATGGAATTCTCTCAGTGGAGGCAGTGGAGACATGGTGGACGGCTTACAAAAAACAGCAGATAGCTCTGTTGAAACCAATGGAACCTCAAAGGATGAAAAGATATCTATGACATCAGTACATAATAGTTCAGAAAACTCGGACACGCGTTCACATATTACACCAGACTCATTAGATATTGATACTACTCCACGCTTTGCTGCCATGCAAGTTAGTTCAGTGTAA
Protein
MSEWKHLITEWLNEYTSLQENEVQSFAAEHEHNHEIATAIFNLFYCEDETVTYNCELENDAQLVQILENVCTQLFSFYRSKEIQLQRFTLQFIPTLIYNYLSSVAQGKKKTYRCIETLLIGIYNFEVVDENGKPKVVSFRLPSLAQSSIYHEPLSLGAQFLTESALRRWEECNTKLVNWGPLPQVDVINAQNRLKIVSALLFVYNRQLSLLPKLSLRHLCIAASRIVTHGFEKKQGNDSKRVPRIAVSPSFLLELIAGAYFAMFNEFYTLALQAVKDVDQRAQYELFPEVMIVTSAVINSLKNNASGQPFDGPMGISVALSPATTTVTMSKSMITNASFRTKKLPDDIPIQAGQATTAESTDVLTSITEEGESETAPIQRGIIARNSKAKLSSFPVLGKKTKDTKDKGNEKKVILKESSKGIWNSLSGGSGDMVDGLQKTADSSVETNGTSKDEKISMTSVHNSSENSDTRSHITPDSLDIDTTPRFAAMQVSSV
Summary
Uniprot
A0A2H1WDE2
A0A2A4JWI8
A0A3S2P573
A0A1E1WE59
A0A194PK45
A0A194RBV5
+ More
A0A212F6X5 A0A2J7QBI2 A0A067RHC1 A0A158ND64 E2AIN9 A0A0K8TRW8 A0A1Q3FUJ3 A0A1Q3FUG0 A0A1L8DMT9 A0A182MP22 A0A182HAW4 A0A1B6EL93 A0A2M4CSC5 A0A1B6EG56 A0A026WWY0 A0A1B6ML45 A0A1B0DMA8 A0A2M4CRV4 A0A2M4BI21 A0A2M4BHC0 A0A2M3Z595 A0A182THL9 A0A2M4AIC9 A0A182KR49 Q7PT63 A0A336M8S9 A0A0V0GAN9 A0A023F8L8 A0A069DV98 A0A224XM80 A0A088A5B8 B0WUG4 F4X7A3 A0A2A3E6K7 A0A023ETS0 A0A0L7QXC4 A0A0A9WNX4 A0A146L6S7 A0A151XHR7 A0A0K8S7B2 A0A151ICE9 R4G8T0 A0A1Y9HEI6 A0A1B0CU24 A0A2M3Z974 Q0IEU8 A0A182PQL8 A0A182WJE3 A0A182JQE4 A0A182NI25 A0A182QAS9 A0A182XYP4 A0A182X3X7 A0A182I342 W5JFR4 A0A1B6CSW0 A0A182VCC3 A0A182JCG4 A0A182FDE9 E2C010 A0A195B0W9 A0A154P0X0 A0A0N7Z8P1 A0A0A9YH21 A0A0K8S784 A0A084WN10 E0VG94 A0A1J1I9F5 W8CDK3 T1PJS0 K7IRS0 A0A1I8MUH8 A0A1W4VCZ3 A0A195DJC3 A0A0A1XKB8 A0A0K8UM74 A0A034VY53 B4MDW1 B3MID4 A0A3B0JNI9 A0A0L0CBA7 A0A1I8Q6L6 B4J3V1 A0A232EVL0 B4GG34 A0A310SV24 Q28XG4 A0A1B0FIU2 A0A1A9WI42 B4KST2 A0A1A9VPR3 A0A1B0BW23 A0A1A9Z7G1
A0A212F6X5 A0A2J7QBI2 A0A067RHC1 A0A158ND64 E2AIN9 A0A0K8TRW8 A0A1Q3FUJ3 A0A1Q3FUG0 A0A1L8DMT9 A0A182MP22 A0A182HAW4 A0A1B6EL93 A0A2M4CSC5 A0A1B6EG56 A0A026WWY0 A0A1B6ML45 A0A1B0DMA8 A0A2M4CRV4 A0A2M4BI21 A0A2M4BHC0 A0A2M3Z595 A0A182THL9 A0A2M4AIC9 A0A182KR49 Q7PT63 A0A336M8S9 A0A0V0GAN9 A0A023F8L8 A0A069DV98 A0A224XM80 A0A088A5B8 B0WUG4 F4X7A3 A0A2A3E6K7 A0A023ETS0 A0A0L7QXC4 A0A0A9WNX4 A0A146L6S7 A0A151XHR7 A0A0K8S7B2 A0A151ICE9 R4G8T0 A0A1Y9HEI6 A0A1B0CU24 A0A2M3Z974 Q0IEU8 A0A182PQL8 A0A182WJE3 A0A182JQE4 A0A182NI25 A0A182QAS9 A0A182XYP4 A0A182X3X7 A0A182I342 W5JFR4 A0A1B6CSW0 A0A182VCC3 A0A182JCG4 A0A182FDE9 E2C010 A0A195B0W9 A0A154P0X0 A0A0N7Z8P1 A0A0A9YH21 A0A0K8S784 A0A084WN10 E0VG94 A0A1J1I9F5 W8CDK3 T1PJS0 K7IRS0 A0A1I8MUH8 A0A1W4VCZ3 A0A195DJC3 A0A0A1XKB8 A0A0K8UM74 A0A034VY53 B4MDW1 B3MID4 A0A3B0JNI9 A0A0L0CBA7 A0A1I8Q6L6 B4J3V1 A0A232EVL0 B4GG34 A0A310SV24 Q28XG4 A0A1B0FIU2 A0A1A9WI42 B4KST2 A0A1A9VPR3 A0A1B0BW23 A0A1A9Z7G1
Pubmed
26354079
22118469
24845553
21347285
20798317
26369729
+ More
26483478 24508170 20966253 12364791 14747013 17210077 25474469 26334808 21719571 24945155 25401762 26823975 17510324 25244985 20920257 23761445 27129103 24438588 20566863 24495485 20075255 25315136 25830018 25348373 17994087 26108605 28648823 15632085
26483478 24508170 20966253 12364791 14747013 17210077 25474469 26334808 21719571 24945155 25401762 26823975 17510324 25244985 20920257 23761445 27129103 24438588 20566863 24495485 20075255 25315136 25830018 25348373 17994087 26108605 28648823 15632085
EMBL
ODYU01007904
SOQ51089.1
NWSH01000473
PCG76179.1
RSAL01000019
RVE52691.1
+ More
GDQN01005913 JAT85141.1 KQ459606 KPI91465.1 KQ460398 KPJ15082.1 AGBW02009960 OWR49491.1 NEVH01016298 PNF25943.1 KK852473 KDR23157.1 ADTU01012423 ADTU01012424 GL439852 EFN66694.1 GDAI01000692 JAI16911.1 GFDL01003775 JAV31270.1 GFDL01003784 JAV31261.1 GFDF01006424 JAV07660.1 AXCM01000608 JXUM01031563 JXUM01031564 JXUM01031565 JXUM01031566 JXUM01031567 JXUM01031568 JXUM01031569 GECZ01031065 JAS38704.1 GGFL01003893 MBW68071.1 GEDC01000427 JAS36871.1 KK107102 EZA59614.1 GEBQ01032084 GEBQ01003321 JAT07893.1 JAT36656.1 AJVK01016712 AJVK01016713 AJVK01016714 AJVK01016715 AJVK01016716 GGFL01003892 MBW68070.1 GGFJ01003317 MBW52458.1 GGFJ01003318 MBW52459.1 GGFM01002924 MBW23675.1 GGFK01007213 MBW40534.1 AAAB01008807 EAA04419.5 UFQT01000550 SSX25183.1 GECL01001096 JAP05028.1 GBBI01000976 JAC17736.1 GBGD01001262 JAC87627.1 GFTR01006724 JAW09702.1 DS232104 EDS34943.1 GL888828 EGI57715.1 KZ288364 PBC26856.1 GAPW01000848 JAC12750.1 KQ414705 KOC63262.1 GBHO01033461 GBHO01033452 JAG10143.1 JAG10152.1 GDHC01015757 JAQ02872.1 KQ982116 KYQ59946.1 GBRD01016660 JAG49167.1 KQ978060 KYM97731.1 ACPB03022602 GAHY01000437 JAA77073.1 AJWK01028337 AJWK01028338 AJWK01028339 AJWK01028340 AJWK01028341 AJWK01028342 GGFM01004284 MBW25035.1 CH477478 EAT40253.1 AXCN02000388 ADMH02001280 ETN63217.1 GEDC01020746 JAS16552.1 GL451712 EFN78706.1 KQ976690 KYM78106.1 KQ434783 KZC04780.1 GDKW01002803 JAI53792.1 GBHO01033470 GBHO01013206 JAG10134.1 JAG30398.1 GBRD01016661 JAG49166.1 ATLV01024547 KE525352 KFB51604.1 DS235133 EEB12400.1 CVRI01000043 CRK96204.1 GAMC01001796 JAC04760.1 KA649001 AFP63630.1 KQ980800 KYN12941.1 GBXI01002987 JAD11305.1 GDHF01024520 JAI27794.1 GAKP01010741 JAC48211.1 CH940662 EDW58726.1 CH902619 EDV36982.1 OUUW01000001 SPP74896.1 JRES01000655 KNC29541.1 CH916367 EDW01534.1 NNAY01001987 OXU22378.1 CH479183 EDW35454.1 KQ759893 OAD62055.1 CM000071 EAL26352.1 CCAG010023811 CH933808 EDW08429.1 JXJN01021578
GDQN01005913 JAT85141.1 KQ459606 KPI91465.1 KQ460398 KPJ15082.1 AGBW02009960 OWR49491.1 NEVH01016298 PNF25943.1 KK852473 KDR23157.1 ADTU01012423 ADTU01012424 GL439852 EFN66694.1 GDAI01000692 JAI16911.1 GFDL01003775 JAV31270.1 GFDL01003784 JAV31261.1 GFDF01006424 JAV07660.1 AXCM01000608 JXUM01031563 JXUM01031564 JXUM01031565 JXUM01031566 JXUM01031567 JXUM01031568 JXUM01031569 GECZ01031065 JAS38704.1 GGFL01003893 MBW68071.1 GEDC01000427 JAS36871.1 KK107102 EZA59614.1 GEBQ01032084 GEBQ01003321 JAT07893.1 JAT36656.1 AJVK01016712 AJVK01016713 AJVK01016714 AJVK01016715 AJVK01016716 GGFL01003892 MBW68070.1 GGFJ01003317 MBW52458.1 GGFJ01003318 MBW52459.1 GGFM01002924 MBW23675.1 GGFK01007213 MBW40534.1 AAAB01008807 EAA04419.5 UFQT01000550 SSX25183.1 GECL01001096 JAP05028.1 GBBI01000976 JAC17736.1 GBGD01001262 JAC87627.1 GFTR01006724 JAW09702.1 DS232104 EDS34943.1 GL888828 EGI57715.1 KZ288364 PBC26856.1 GAPW01000848 JAC12750.1 KQ414705 KOC63262.1 GBHO01033461 GBHO01033452 JAG10143.1 JAG10152.1 GDHC01015757 JAQ02872.1 KQ982116 KYQ59946.1 GBRD01016660 JAG49167.1 KQ978060 KYM97731.1 ACPB03022602 GAHY01000437 JAA77073.1 AJWK01028337 AJWK01028338 AJWK01028339 AJWK01028340 AJWK01028341 AJWK01028342 GGFM01004284 MBW25035.1 CH477478 EAT40253.1 AXCN02000388 ADMH02001280 ETN63217.1 GEDC01020746 JAS16552.1 GL451712 EFN78706.1 KQ976690 KYM78106.1 KQ434783 KZC04780.1 GDKW01002803 JAI53792.1 GBHO01033470 GBHO01013206 JAG10134.1 JAG30398.1 GBRD01016661 JAG49166.1 ATLV01024547 KE525352 KFB51604.1 DS235133 EEB12400.1 CVRI01000043 CRK96204.1 GAMC01001796 JAC04760.1 KA649001 AFP63630.1 KQ980800 KYN12941.1 GBXI01002987 JAD11305.1 GDHF01024520 JAI27794.1 GAKP01010741 JAC48211.1 CH940662 EDW58726.1 CH902619 EDV36982.1 OUUW01000001 SPP74896.1 JRES01000655 KNC29541.1 CH916367 EDW01534.1 NNAY01001987 OXU22378.1 CH479183 EDW35454.1 KQ759893 OAD62055.1 CM000071 EAL26352.1 CCAG010023811 CH933808 EDW08429.1 JXJN01021578
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
UP000235965
+ More
UP000027135 UP000005205 UP000000311 UP000075883 UP000069940 UP000053097 UP000092462 UP000075902 UP000075882 UP000007062 UP000005203 UP000002320 UP000007755 UP000242457 UP000053825 UP000075809 UP000078542 UP000015103 UP000075900 UP000092461 UP000008820 UP000075885 UP000075920 UP000075881 UP000075884 UP000075886 UP000076408 UP000000673 UP000075903 UP000075880 UP000069272 UP000008237 UP000078540 UP000076502 UP000030765 UP000009046 UP000183832 UP000002358 UP000095301 UP000192221 UP000078492 UP000008792 UP000007801 UP000268350 UP000037069 UP000095300 UP000001070 UP000215335 UP000008744 UP000001819 UP000092444 UP000091820 UP000009192 UP000078200 UP000092460 UP000092445
UP000027135 UP000005205 UP000000311 UP000075883 UP000069940 UP000053097 UP000092462 UP000075902 UP000075882 UP000007062 UP000005203 UP000002320 UP000007755 UP000242457 UP000053825 UP000075809 UP000078542 UP000015103 UP000075900 UP000092461 UP000008820 UP000075885 UP000075920 UP000075881 UP000075884 UP000075886 UP000076408 UP000000673 UP000075903 UP000075880 UP000069272 UP000008237 UP000078540 UP000076502 UP000030765 UP000009046 UP000183832 UP000002358 UP000095301 UP000192221 UP000078492 UP000008792 UP000007801 UP000268350 UP000037069 UP000095300 UP000001070 UP000215335 UP000008744 UP000001819 UP000092444 UP000091820 UP000009192 UP000078200 UP000092460 UP000092445
Pfam
PF09790 Hyccin
Interpro
IPR018619
Hyccin
ProteinModelPortal
A0A2H1WDE2
A0A2A4JWI8
A0A3S2P573
A0A1E1WE59
A0A194PK45
A0A194RBV5
+ More
A0A212F6X5 A0A2J7QBI2 A0A067RHC1 A0A158ND64 E2AIN9 A0A0K8TRW8 A0A1Q3FUJ3 A0A1Q3FUG0 A0A1L8DMT9 A0A182MP22 A0A182HAW4 A0A1B6EL93 A0A2M4CSC5 A0A1B6EG56 A0A026WWY0 A0A1B6ML45 A0A1B0DMA8 A0A2M4CRV4 A0A2M4BI21 A0A2M4BHC0 A0A2M3Z595 A0A182THL9 A0A2M4AIC9 A0A182KR49 Q7PT63 A0A336M8S9 A0A0V0GAN9 A0A023F8L8 A0A069DV98 A0A224XM80 A0A088A5B8 B0WUG4 F4X7A3 A0A2A3E6K7 A0A023ETS0 A0A0L7QXC4 A0A0A9WNX4 A0A146L6S7 A0A151XHR7 A0A0K8S7B2 A0A151ICE9 R4G8T0 A0A1Y9HEI6 A0A1B0CU24 A0A2M3Z974 Q0IEU8 A0A182PQL8 A0A182WJE3 A0A182JQE4 A0A182NI25 A0A182QAS9 A0A182XYP4 A0A182X3X7 A0A182I342 W5JFR4 A0A1B6CSW0 A0A182VCC3 A0A182JCG4 A0A182FDE9 E2C010 A0A195B0W9 A0A154P0X0 A0A0N7Z8P1 A0A0A9YH21 A0A0K8S784 A0A084WN10 E0VG94 A0A1J1I9F5 W8CDK3 T1PJS0 K7IRS0 A0A1I8MUH8 A0A1W4VCZ3 A0A195DJC3 A0A0A1XKB8 A0A0K8UM74 A0A034VY53 B4MDW1 B3MID4 A0A3B0JNI9 A0A0L0CBA7 A0A1I8Q6L6 B4J3V1 A0A232EVL0 B4GG34 A0A310SV24 Q28XG4 A0A1B0FIU2 A0A1A9WI42 B4KST2 A0A1A9VPR3 A0A1B0BW23 A0A1A9Z7G1
A0A212F6X5 A0A2J7QBI2 A0A067RHC1 A0A158ND64 E2AIN9 A0A0K8TRW8 A0A1Q3FUJ3 A0A1Q3FUG0 A0A1L8DMT9 A0A182MP22 A0A182HAW4 A0A1B6EL93 A0A2M4CSC5 A0A1B6EG56 A0A026WWY0 A0A1B6ML45 A0A1B0DMA8 A0A2M4CRV4 A0A2M4BI21 A0A2M4BHC0 A0A2M3Z595 A0A182THL9 A0A2M4AIC9 A0A182KR49 Q7PT63 A0A336M8S9 A0A0V0GAN9 A0A023F8L8 A0A069DV98 A0A224XM80 A0A088A5B8 B0WUG4 F4X7A3 A0A2A3E6K7 A0A023ETS0 A0A0L7QXC4 A0A0A9WNX4 A0A146L6S7 A0A151XHR7 A0A0K8S7B2 A0A151ICE9 R4G8T0 A0A1Y9HEI6 A0A1B0CU24 A0A2M3Z974 Q0IEU8 A0A182PQL8 A0A182WJE3 A0A182JQE4 A0A182NI25 A0A182QAS9 A0A182XYP4 A0A182X3X7 A0A182I342 W5JFR4 A0A1B6CSW0 A0A182VCC3 A0A182JCG4 A0A182FDE9 E2C010 A0A195B0W9 A0A154P0X0 A0A0N7Z8P1 A0A0A9YH21 A0A0K8S784 A0A084WN10 E0VG94 A0A1J1I9F5 W8CDK3 T1PJS0 K7IRS0 A0A1I8MUH8 A0A1W4VCZ3 A0A195DJC3 A0A0A1XKB8 A0A0K8UM74 A0A034VY53 B4MDW1 B3MID4 A0A3B0JNI9 A0A0L0CBA7 A0A1I8Q6L6 B4J3V1 A0A232EVL0 B4GG34 A0A310SV24 Q28XG4 A0A1B0FIU2 A0A1A9WI42 B4KST2 A0A1A9VPR3 A0A1B0BW23 A0A1A9Z7G1
PDB
5DSE
E-value=9.36625e-52,
Score=515
Ontologies
PANTHER
Topology
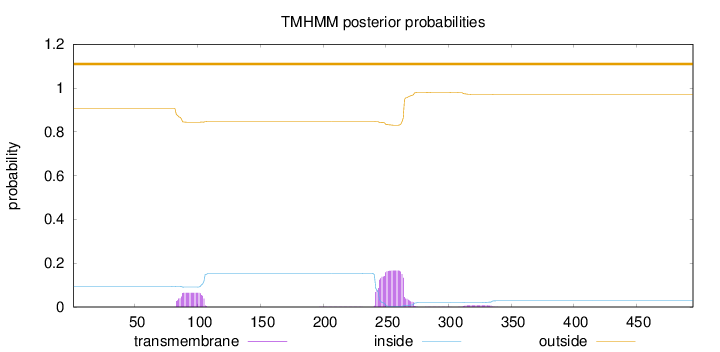
Length:
495
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.10454
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09393
outside
1 - 495
Population Genetic Test Statistics
Pi
240.853365
Theta
226.377306
Tajima's D
-1.324833
CLR
0.901302
CSRT
0.0839958002099895
Interpretation
Possibly Positive selection
