Gene
KWMTBOMO05704
Annotation
PREDICTED:_sodium-coupled_monocarboxylate_transporter_1_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.784
Sequence
CDS
ATGATGATAACGGGTCTAAATACATATCTAGCAGTATCATTGGTGTATGTCGTATGCATTTTCTACGCATCACAGGGTGGCATGAAAGCAGTCATCATGACAGACTCATTTCAGTCAGCAGTATTGGTAGGATCTCTAGCCACGGTGTTGGCGATGGGTGCTTCGAGGGTTGGAGGCTTAGAAGCAGTTTGGAAACACGCTAGCCTCACGGACAGACTCCACTTTTTTGAGTAA
Protein
MMITGLNTYLAVSLVYVVCIFYASQGGMKAVIMTDSFQSAVLVGSLATVLAMGASRVGGLEAVWKHASLTDRLHFFE
Summary
Similarity
Belongs to the sodium:solute symporter (SSF) (TC 2.A.21) family.
Uniprot
A0A194RC90
A0A2H1VGN3
A0A2A4JVV6
A0A212F6Y7
A0A3S2LFK1
H9JBG5
+ More
E2AIX1 A0A0M8ZY87 A0A151X8Z7 A0A195CL10 E9JAQ7 A0A195DFZ3 A0A026X4I2 A0A3L8D8E8 F4X3I9 A0A232EIL3 A0A195EXC1 A0A067QP53 A0A195BN99 A0A0J7KVI8 A0A0L7R0N4 E2B5H5 A0A0C9RRN6 A0A158NRN5 A0A310SJQ4 A0A1B6DS39 A0A087ZNV2 A0A154P042 A0A2J7QJS2 A0A2J7QJT7 A0A1Y1MH80 A0A1Y1MCD1 A0A139WI01 A0A139WI19 A0A1B6M044 A0A2R7WYH9 A0A1B6IF30 A0A2A3ENK4 A0A023F6H1 A0A1I8PX36 A0A1B6F543 A0A1B6M0L6 A0A1B6F1B8 A0A1B6J094 A0A1B6CGF1 A0A0L7RCU1 B4HW29 A0A0K8V6R8 B4PE95 A0A226E1U3 B3NEY0 N6TA95 A0A226EL08 A0A0T6AZE2 Q961Q9 B4QN09 A0A0A9ZFM9 A0A146LXE4 A0A0A9ZC26 A0A226DZ22 A0A1A9W4N0 A0A1W4VVB7 B3M780 B4GS39 Q2M053 A0A0K8VCB8 A0A0K8U2A4 A0A226EQQ5 A0A1B0BZ70 A0A1A9YLR2 A0A3B0JRX9 A0A1B0G5R2 A0A1A9ZMT8 B4N328 A0A0M8ZM94 E2AWG6 A0A1W4WGZ1 A0A084WDJ5 A0A1A9UXU1 T1HZD6 A0A182PZH6 B0VZN2 A0A1B0CPY2 Q7Q6K4 A0A2C9H5N5 A0A2C9H889 A0A2C9GQ50 A0A3L8DIG3 A0A088AH95 A0A026X262 A0A3L8DFP4 A0A195BBB9 A0A3M6V2I6 A0A2J7PCD1 A0A2J7PCD0 A0A195BZT4 A0A154PA03 A0A139WI69 D6WJW9
E2AIX1 A0A0M8ZY87 A0A151X8Z7 A0A195CL10 E9JAQ7 A0A195DFZ3 A0A026X4I2 A0A3L8D8E8 F4X3I9 A0A232EIL3 A0A195EXC1 A0A067QP53 A0A195BN99 A0A0J7KVI8 A0A0L7R0N4 E2B5H5 A0A0C9RRN6 A0A158NRN5 A0A310SJQ4 A0A1B6DS39 A0A087ZNV2 A0A154P042 A0A2J7QJS2 A0A2J7QJT7 A0A1Y1MH80 A0A1Y1MCD1 A0A139WI01 A0A139WI19 A0A1B6M044 A0A2R7WYH9 A0A1B6IF30 A0A2A3ENK4 A0A023F6H1 A0A1I8PX36 A0A1B6F543 A0A1B6M0L6 A0A1B6F1B8 A0A1B6J094 A0A1B6CGF1 A0A0L7RCU1 B4HW29 A0A0K8V6R8 B4PE95 A0A226E1U3 B3NEY0 N6TA95 A0A226EL08 A0A0T6AZE2 Q961Q9 B4QN09 A0A0A9ZFM9 A0A146LXE4 A0A0A9ZC26 A0A226DZ22 A0A1A9W4N0 A0A1W4VVB7 B3M780 B4GS39 Q2M053 A0A0K8VCB8 A0A0K8U2A4 A0A226EQQ5 A0A1B0BZ70 A0A1A9YLR2 A0A3B0JRX9 A0A1B0G5R2 A0A1A9ZMT8 B4N328 A0A0M8ZM94 E2AWG6 A0A1W4WGZ1 A0A084WDJ5 A0A1A9UXU1 T1HZD6 A0A182PZH6 B0VZN2 A0A1B0CPY2 Q7Q6K4 A0A2C9H5N5 A0A2C9H889 A0A2C9GQ50 A0A3L8DIG3 A0A088AH95 A0A026X262 A0A3L8DFP4 A0A195BBB9 A0A3M6V2I6 A0A2J7PCD1 A0A2J7PCD0 A0A195BZT4 A0A154PA03 A0A139WI69 D6WJW9
Pubmed
26354079
22118469
19121390
20798317
21282665
24508170
+ More
30249741 21719571 28648823 24845553 21347285 28004739 18362917 19820115 25474469 17994087 17550304 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25401762 26823975 15632085 24438588 12364791 30382153
30249741 21719571 28648823 24845553 21347285 28004739 18362917 19820115 25474469 17994087 17550304 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25401762 26823975 15632085 24438588 12364791 30382153
EMBL
KQ460398
KPJ15084.1
ODYU01002461
SOQ39976.1
NWSH01000473
PCG76177.1
+ More
AGBW02009960 OWR49493.1 RSAL01000019 RVE52689.1 BABH01030875 BABH01030876 GL439905 EFN66650.1 KQ435798 KOX73367.1 KQ982409 KYQ56790.1 KQ977649 KYN00769.1 GL770679 EFZ10023.1 KQ980886 KYN11813.1 KK107021 EZA62319.1 QOIP01000011 RLU16561.1 GL888613 EGI59017.1 NNAY01004195 OXU18217.1 KQ981931 KYN32796.1 KK853117 KDR11160.1 KQ976439 KYM86676.1 LBMM01002853 KMQ94279.1 KQ414670 KOC64351.1 GL445827 EFN89061.1 GBYB01011215 JAG80982.1 ADTU01024213 ADTU01024214 KQ762329 OAD55963.1 GEDC01008812 JAS28486.1 KQ434781 KZC04500.1 NEVH01013549 PNF28843.1 PNF28844.1 GEZM01036474 GEZM01036473 JAV82667.1 GEZM01036472 GEZM01036471 JAV82668.1 KQ971342 KYB27603.1 KYB27602.1 GEBQ01010674 JAT29303.1 KK855873 PTY24210.1 GECU01022160 JAS85546.1 KZ288204 PBC33280.1 GBBI01001874 JAC16838.1 GECZ01024558 JAS45211.1 GEBQ01010509 JAT29468.1 GECZ01025673 JAS44096.1 GECU01015092 JAS92614.1 GEDC01024722 JAS12576.1 KQ414614 KOC68807.1 CH480817 EDW50144.1 GDHF01017687 JAI34627.1 CM000159 EDW92933.1 LNIX01000009 OXA50446.1 CH954178 EDV50253.1 APGK01037934 KB740949 ENN77189.1 LNIX01000003 OXA58385.1 LJIG01022449 KRT80547.1 AE014296 AY051427 AAF47595.3 AAK92851.1 CM000363 CM002912 EDX08891.1 KMY96988.1 GBHO01038683 GBHO01038678 GBHO01036758 GBHO01036014 GBHO01033555 GBHO01033552 GBHO01005171 GBHO01004442 GBHO01000800 GBHO01000798 GBHO01000793 JAG04921.1 JAG04926.1 JAG06846.1 JAG07590.1 JAG10049.1 JAG10052.1 JAG38433.1 JAG39162.1 JAG42804.1 JAG42806.1 JAG42811.1 GDHC01013951 GDHC01007287 JAQ04678.1 JAQ11342.1 GBHO01038679 GBHO01038677 GBHO01038676 GBHO01038673 GBHO01038672 GBHO01035996 GBHO01033551 GBHO01019355 GBHO01000802 GBHO01000797 GBHO01000792 JAG04925.1 JAG04927.1 JAG04928.1 JAG04931.1 JAG04932.1 JAG07608.1 JAG10053.1 JAG24249.1 JAG42802.1 JAG42807.1 JAG42812.1 OXA50044.1 CH902618 EDV38741.1 CH479188 EDW40574.1 CH379069 EAL31083.2 GDHF01015827 JAI36487.1 GDHF01031440 JAI20874.1 LNIX01000002 OXA58926.1 JXJN01023005 JXJN01023006 OUUW01000009 SPP84877.1 CCAG010018755 CCAG010018756 CH964062 EDW78767.1 KQ438551 KOX67190.1 GL443286 EFN62266.1 ATLV01023033 KE525339 KFB48289.1 ACPB03008662 AXCN02000442 AXCN02000443 DS231815 EDS35124.1 AJWK01022836 AJWK01022837 AJWK01022838 AJWK01022839 AJWK01022840 AAAB01008960 EAA11888.4 APCN01000142 QOIP01000008 RLU19628.1 KK107031 EZA62128.1 RLU19285.1 KQ976528 KYM81828.1 RCHS01000245 RMX60097.1 NEVH01027063 PNF13988.1 PNF13986.1 KQ978457 KYM93850.1 KQ434856 KZC08682.1 KYB27604.1 EFA03651.2
AGBW02009960 OWR49493.1 RSAL01000019 RVE52689.1 BABH01030875 BABH01030876 GL439905 EFN66650.1 KQ435798 KOX73367.1 KQ982409 KYQ56790.1 KQ977649 KYN00769.1 GL770679 EFZ10023.1 KQ980886 KYN11813.1 KK107021 EZA62319.1 QOIP01000011 RLU16561.1 GL888613 EGI59017.1 NNAY01004195 OXU18217.1 KQ981931 KYN32796.1 KK853117 KDR11160.1 KQ976439 KYM86676.1 LBMM01002853 KMQ94279.1 KQ414670 KOC64351.1 GL445827 EFN89061.1 GBYB01011215 JAG80982.1 ADTU01024213 ADTU01024214 KQ762329 OAD55963.1 GEDC01008812 JAS28486.1 KQ434781 KZC04500.1 NEVH01013549 PNF28843.1 PNF28844.1 GEZM01036474 GEZM01036473 JAV82667.1 GEZM01036472 GEZM01036471 JAV82668.1 KQ971342 KYB27603.1 KYB27602.1 GEBQ01010674 JAT29303.1 KK855873 PTY24210.1 GECU01022160 JAS85546.1 KZ288204 PBC33280.1 GBBI01001874 JAC16838.1 GECZ01024558 JAS45211.1 GEBQ01010509 JAT29468.1 GECZ01025673 JAS44096.1 GECU01015092 JAS92614.1 GEDC01024722 JAS12576.1 KQ414614 KOC68807.1 CH480817 EDW50144.1 GDHF01017687 JAI34627.1 CM000159 EDW92933.1 LNIX01000009 OXA50446.1 CH954178 EDV50253.1 APGK01037934 KB740949 ENN77189.1 LNIX01000003 OXA58385.1 LJIG01022449 KRT80547.1 AE014296 AY051427 AAF47595.3 AAK92851.1 CM000363 CM002912 EDX08891.1 KMY96988.1 GBHO01038683 GBHO01038678 GBHO01036758 GBHO01036014 GBHO01033555 GBHO01033552 GBHO01005171 GBHO01004442 GBHO01000800 GBHO01000798 GBHO01000793 JAG04921.1 JAG04926.1 JAG06846.1 JAG07590.1 JAG10049.1 JAG10052.1 JAG38433.1 JAG39162.1 JAG42804.1 JAG42806.1 JAG42811.1 GDHC01013951 GDHC01007287 JAQ04678.1 JAQ11342.1 GBHO01038679 GBHO01038677 GBHO01038676 GBHO01038673 GBHO01038672 GBHO01035996 GBHO01033551 GBHO01019355 GBHO01000802 GBHO01000797 GBHO01000792 JAG04925.1 JAG04927.1 JAG04928.1 JAG04931.1 JAG04932.1 JAG07608.1 JAG10053.1 JAG24249.1 JAG42802.1 JAG42807.1 JAG42812.1 OXA50044.1 CH902618 EDV38741.1 CH479188 EDW40574.1 CH379069 EAL31083.2 GDHF01015827 JAI36487.1 GDHF01031440 JAI20874.1 LNIX01000002 OXA58926.1 JXJN01023005 JXJN01023006 OUUW01000009 SPP84877.1 CCAG010018755 CCAG010018756 CH964062 EDW78767.1 KQ438551 KOX67190.1 GL443286 EFN62266.1 ATLV01023033 KE525339 KFB48289.1 ACPB03008662 AXCN02000442 AXCN02000443 DS231815 EDS35124.1 AJWK01022836 AJWK01022837 AJWK01022838 AJWK01022839 AJWK01022840 AAAB01008960 EAA11888.4 APCN01000142 QOIP01000008 RLU19628.1 KK107031 EZA62128.1 RLU19285.1 KQ976528 KYM81828.1 RCHS01000245 RMX60097.1 NEVH01027063 PNF13988.1 PNF13986.1 KQ978457 KYM93850.1 KQ434856 KZC08682.1 KYB27604.1 EFA03651.2
Proteomes
UP000053240
UP000218220
UP000007151
UP000283053
UP000005204
UP000000311
+ More
UP000053105 UP000075809 UP000078542 UP000078492 UP000053097 UP000279307 UP000007755 UP000215335 UP000078541 UP000027135 UP000078540 UP000036403 UP000053825 UP000008237 UP000005205 UP000005203 UP000076502 UP000235965 UP000007266 UP000242457 UP000095300 UP000001292 UP000002282 UP000198287 UP000008711 UP000019118 UP000000803 UP000000304 UP000091820 UP000192221 UP000007801 UP000008744 UP000001819 UP000092460 UP000092443 UP000268350 UP000092444 UP000092445 UP000007798 UP000192223 UP000030765 UP000078200 UP000015103 UP000075886 UP000002320 UP000092461 UP000007062 UP000075903 UP000076407 UP000075840 UP000275408
UP000053105 UP000075809 UP000078542 UP000078492 UP000053097 UP000279307 UP000007755 UP000215335 UP000078541 UP000027135 UP000078540 UP000036403 UP000053825 UP000008237 UP000005205 UP000005203 UP000076502 UP000235965 UP000007266 UP000242457 UP000095300 UP000001292 UP000002282 UP000198287 UP000008711 UP000019118 UP000000803 UP000000304 UP000091820 UP000192221 UP000007801 UP000008744 UP000001819 UP000092460 UP000092443 UP000268350 UP000092444 UP000092445 UP000007798 UP000192223 UP000030765 UP000078200 UP000015103 UP000075886 UP000002320 UP000092461 UP000007062 UP000075903 UP000076407 UP000075840 UP000275408
Interpro
Gene 3D
ProteinModelPortal
A0A194RC90
A0A2H1VGN3
A0A2A4JVV6
A0A212F6Y7
A0A3S2LFK1
H9JBG5
+ More
E2AIX1 A0A0M8ZY87 A0A151X8Z7 A0A195CL10 E9JAQ7 A0A195DFZ3 A0A026X4I2 A0A3L8D8E8 F4X3I9 A0A232EIL3 A0A195EXC1 A0A067QP53 A0A195BN99 A0A0J7KVI8 A0A0L7R0N4 E2B5H5 A0A0C9RRN6 A0A158NRN5 A0A310SJQ4 A0A1B6DS39 A0A087ZNV2 A0A154P042 A0A2J7QJS2 A0A2J7QJT7 A0A1Y1MH80 A0A1Y1MCD1 A0A139WI01 A0A139WI19 A0A1B6M044 A0A2R7WYH9 A0A1B6IF30 A0A2A3ENK4 A0A023F6H1 A0A1I8PX36 A0A1B6F543 A0A1B6M0L6 A0A1B6F1B8 A0A1B6J094 A0A1B6CGF1 A0A0L7RCU1 B4HW29 A0A0K8V6R8 B4PE95 A0A226E1U3 B3NEY0 N6TA95 A0A226EL08 A0A0T6AZE2 Q961Q9 B4QN09 A0A0A9ZFM9 A0A146LXE4 A0A0A9ZC26 A0A226DZ22 A0A1A9W4N0 A0A1W4VVB7 B3M780 B4GS39 Q2M053 A0A0K8VCB8 A0A0K8U2A4 A0A226EQQ5 A0A1B0BZ70 A0A1A9YLR2 A0A3B0JRX9 A0A1B0G5R2 A0A1A9ZMT8 B4N328 A0A0M8ZM94 E2AWG6 A0A1W4WGZ1 A0A084WDJ5 A0A1A9UXU1 T1HZD6 A0A182PZH6 B0VZN2 A0A1B0CPY2 Q7Q6K4 A0A2C9H5N5 A0A2C9H889 A0A2C9GQ50 A0A3L8DIG3 A0A088AH95 A0A026X262 A0A3L8DFP4 A0A195BBB9 A0A3M6V2I6 A0A2J7PCD1 A0A2J7PCD0 A0A195BZT4 A0A154PA03 A0A139WI69 D6WJW9
E2AIX1 A0A0M8ZY87 A0A151X8Z7 A0A195CL10 E9JAQ7 A0A195DFZ3 A0A026X4I2 A0A3L8D8E8 F4X3I9 A0A232EIL3 A0A195EXC1 A0A067QP53 A0A195BN99 A0A0J7KVI8 A0A0L7R0N4 E2B5H5 A0A0C9RRN6 A0A158NRN5 A0A310SJQ4 A0A1B6DS39 A0A087ZNV2 A0A154P042 A0A2J7QJS2 A0A2J7QJT7 A0A1Y1MH80 A0A1Y1MCD1 A0A139WI01 A0A139WI19 A0A1B6M044 A0A2R7WYH9 A0A1B6IF30 A0A2A3ENK4 A0A023F6H1 A0A1I8PX36 A0A1B6F543 A0A1B6M0L6 A0A1B6F1B8 A0A1B6J094 A0A1B6CGF1 A0A0L7RCU1 B4HW29 A0A0K8V6R8 B4PE95 A0A226E1U3 B3NEY0 N6TA95 A0A226EL08 A0A0T6AZE2 Q961Q9 B4QN09 A0A0A9ZFM9 A0A146LXE4 A0A0A9ZC26 A0A226DZ22 A0A1A9W4N0 A0A1W4VVB7 B3M780 B4GS39 Q2M053 A0A0K8VCB8 A0A0K8U2A4 A0A226EQQ5 A0A1B0BZ70 A0A1A9YLR2 A0A3B0JRX9 A0A1B0G5R2 A0A1A9ZMT8 B4N328 A0A0M8ZM94 E2AWG6 A0A1W4WGZ1 A0A084WDJ5 A0A1A9UXU1 T1HZD6 A0A182PZH6 B0VZN2 A0A1B0CPY2 Q7Q6K4 A0A2C9H5N5 A0A2C9H889 A0A2C9GQ50 A0A3L8DIG3 A0A088AH95 A0A026X262 A0A3L8DFP4 A0A195BBB9 A0A3M6V2I6 A0A2J7PCD1 A0A2J7PCD0 A0A195BZT4 A0A154PA03 A0A139WI69 D6WJW9
PDB
5NVA
E-value=0.0798893,
Score=77
Ontologies
GO
Topology
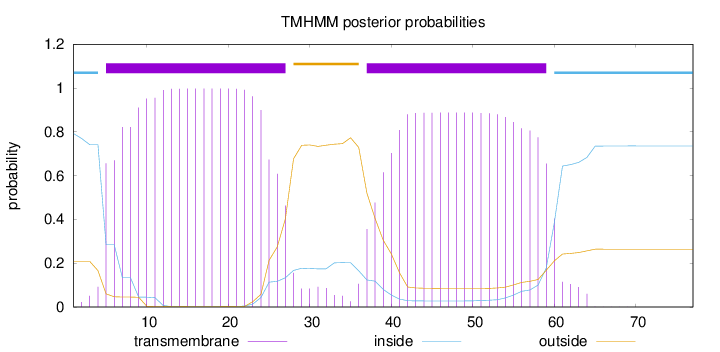
Length:
77
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
40.38098
Exp number, first 60 AAs:
40.00718
Total prob of N-in:
0.79205
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 36
TMhelix
37 - 59
inside
60 - 77
Population Genetic Test Statistics
Pi
288.718118
Theta
220.073406
Tajima's D
0.88858
CLR
0
CSRT
0.628168591570421
Interpretation
Uncertain
