Gene
KWMTBOMO05700 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006859
Annotation
mitochondrial_NADH:ubiquinone_oxidoreductase_ESSS_subunit?_putative_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.476
Sequence
CDS
ATGGCCGCTTTGATAAAACTACGCCATATGCCCTTAGTGCAAAGAAGCGTTTGGAATCATTTCAACAAAGTAAGTCGGAATATTTCCACCTCTAAAAAGAACAGCGACACCGCCACAACTGCTTGTGAGACCAAAACAGAAGATAAGAACTGGGTTAGCTACGGTTTTGACTATACGAGCAAAGAAGAAGACACTAATGCCCATCACGCCACCTTTTTCTTTTCTGTAACATTATGTATTGTTTTTGGTGGTGTCTGTTGGGCCTATTCTCCCGATGTACACATGAGGGACTGGGCTCAGCGTGAAGCATTTTTGGAATTAAGACGTCGTGAAAAAGCAGGTCTTCCACCGATCAGTCCCAACTTTATTGATCCCAAACTTATAAAGCTGCCTGATGAATCCGAGTTGTGTGATGTTGAAATAATCATCTAA
Protein
MAALIKLRHMPLVQRSVWNHFNKVSRNISTSKKNSDTATTACETKTEDKNWVSYGFDYTSKEEDTNAHHATFFFSVTLCIVFGGVCWAYSPDVHMRDWAQREAFLELRRREKAGLPPISPNFIDPKLIKLPDESELCDVEIII
Summary
Uniprot
H9JBG4
I4DK86
A0A194RBW0
A0A0L7LTQ9
A0A212F6X2
A0A2A4JX06
+ More
A0A2H1VIK0 S4PGP5 A0A067RH50 A0A182HAJ3 A0A2J7Q8H0 A0A0K8TPL1 A0A2M3Z6C9 A0A2M4C2E0 W5JJY1 A0A2M4C2D9 T1DFD6 A0A182VAH5 B4QTW2 A0A2M4AJS5 A0A0C9QRN3 A0A034VJH8 A0A182IK98 A0A182KIG7 A0A1L8EF04 A0A0L0CB83 A0A336L4P8 A0A182I347 A0A023EFY4 A0A084VD60 B4ILV7 A0A1I8MQN3 B4PMQ8 A0A182PQL2 A0A182TDQ3 A0A1I8P0Z4 A0A1Y9J0G2 A0A182KR52 A7URF1 A0A182XYQ0 B3P382 Q0IEU1 A0A182Q0I3 A0A1A9YUG3 B4MBW3 A0A182FDE3 A0A1Q3FYR9 W8CE99 J9K6Q7 Q9V3L7 A0A182MS06 B3LVB5 A0A1A9UJX9 B4K5Z1 A0A1Y9IWE2 A0A1W4UAV7 A0A1B0C382 A0A1B0DJQ5 A0A1A9W6P5 V5GV76 B4JYK5 A0A182NI19 B4NHZ0 Q29BT2 A0A1L8E937 B4GPP1 A0A182R7B6 A0A0N7ZDC2 U5EES5 A0A0A9WSU8 A0A1B0CV98 A0A1A9XN11 A0A1L8DZQ3 A0A1J1IB52 A0A2P8YJT3 A0A151XDY3 A0A1W4WQC4 T1I5A7 A0A069DNU3 R4WTB3 N6SYZ1 A0A1S3DAV6 A0A195DW01 A0A1B6CJL6 A0A1Y1JWN2 A0A0J7KT58 A0A2T7NUR1 A0A0M9ACY3 A0A158NWV3 A0A2A3E6R9 A0A195F5A2 F4X4F9 A0A2R7VVS7 A0A0L7QXD1 T1ERM6 E2BBS0 E1ZWY6 A0A154NZ72
A0A2H1VIK0 S4PGP5 A0A067RH50 A0A182HAJ3 A0A2J7Q8H0 A0A0K8TPL1 A0A2M3Z6C9 A0A2M4C2E0 W5JJY1 A0A2M4C2D9 T1DFD6 A0A182VAH5 B4QTW2 A0A2M4AJS5 A0A0C9QRN3 A0A034VJH8 A0A182IK98 A0A182KIG7 A0A1L8EF04 A0A0L0CB83 A0A336L4P8 A0A182I347 A0A023EFY4 A0A084VD60 B4ILV7 A0A1I8MQN3 B4PMQ8 A0A182PQL2 A0A182TDQ3 A0A1I8P0Z4 A0A1Y9J0G2 A0A182KR52 A7URF1 A0A182XYQ0 B3P382 Q0IEU1 A0A182Q0I3 A0A1A9YUG3 B4MBW3 A0A182FDE3 A0A1Q3FYR9 W8CE99 J9K6Q7 Q9V3L7 A0A182MS06 B3LVB5 A0A1A9UJX9 B4K5Z1 A0A1Y9IWE2 A0A1W4UAV7 A0A1B0C382 A0A1B0DJQ5 A0A1A9W6P5 V5GV76 B4JYK5 A0A182NI19 B4NHZ0 Q29BT2 A0A1L8E937 B4GPP1 A0A182R7B6 A0A0N7ZDC2 U5EES5 A0A0A9WSU8 A0A1B0CV98 A0A1A9XN11 A0A1L8DZQ3 A0A1J1IB52 A0A2P8YJT3 A0A151XDY3 A0A1W4WQC4 T1I5A7 A0A069DNU3 R4WTB3 N6SYZ1 A0A1S3DAV6 A0A195DW01 A0A1B6CJL6 A0A1Y1JWN2 A0A0J7KT58 A0A2T7NUR1 A0A0M9ACY3 A0A158NWV3 A0A2A3E6R9 A0A195F5A2 F4X4F9 A0A2R7VVS7 A0A0L7QXD1 T1ERM6 E2BBS0 E1ZWY6 A0A154NZ72
Pubmed
19121390
22651552
26354079
26227816
22118469
23622113
+ More
24845553 26483478 26369729 20920257 23761445 17994087 25348373 26108605 24945155 24438588 25315136 17550304 20966253 12364791 14747013 17210077 25244985 17510324 24495485 10858562 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25401762 26823975 29403074 26334808 23691247 23537049 28004739 21347285 21719571 23254933 20798317
24845553 26483478 26369729 20920257 23761445 17994087 25348373 26108605 24945155 24438588 25315136 17550304 20966253 12364791 14747013 17210077 25244985 17510324 24495485 10858562 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25401762 26823975 29403074 26334808 23691247 23537049 28004739 21347285 21719571 23254933 20798317
EMBL
BABH01030874
AK401704
KQ459606
BAM18326.1
KPI91471.1
KQ460398
+ More
KPJ15087.1 JTDY01000103 KOB78850.1 AGBW02009960 OWR49496.1 NWSH01000473 PCG76174.1 ODYU01002761 SOQ40658.1 GAIX01006120 JAA86440.1 KK852473 KDR23102.1 JXUM01122788 KQ566643 KXJ69952.1 NEVH01016952 PNF24875.1 GDAI01001527 JAI16076.1 GGFM01003328 MBW24079.1 GGFJ01010314 MBW59455.1 ADMH02001280 ETN63210.1 GGFJ01010313 MBW59454.1 GAMD01003176 JAA98414.1 CM000364 EDX12385.1 GGFK01007693 MBW41014.1 GBYB01003292 JAG73059.1 GAKP01015511 GAKP01015507 GAKP01015504 GAKP01015500 GAKP01015496 GAKP01015492 GAKP01015483 GAKP01015480 JAC43456.1 GFDG01001630 JAV17169.1 JRES01000657 KNC29480.1 UFQS01001990 UFQT01001990 SSX12886.1 SSX32328.1 APCN01000197 GAPW01005346 JAC08252.1 ATLV01011185 KE524650 KFB35904.1 CH480892 EDW55433.1 CM000160 EDW95978.1 AAAB01008807 EDO64594.1 CH954181 EDV48534.1 CH477478 EAT40260.1 AXCN02000387 CCAG010023983 CH940656 EDW58584.1 GFDL01002375 JAV32670.1 GAMC01000566 JAC05990.1 ABLF02017094 AF170829 AE014297 AY047546 KX531571 AAD49739.1 AAF55625.1 AAK77278.1 ANY27381.1 AXCM01000608 CH902617 EDV43647.1 CH933806 EDW16228.1 JXJN01024853 AJVK01035127 GALX01002889 JAB65577.1 CH916377 EDV90767.1 CH964272 EDW83640.1 CM000070 EAL26915.1 GFDG01003674 JAV15125.1 CH479186 EDW39124.1 GDRN01045575 JAI66840.1 GANO01004185 JAB55686.1 GBHO01033108 GBRD01014320 GDHC01012365 JAG10496.1 JAG51506.1 JAQ06264.1 AJWK01030327 GFDF01002131 JAV11953.1 CVRI01000043 CRK96206.1 PYGN01000545 PSN44502.1 KQ982254 KYQ58586.1 ACPB03022603 GBGD01003394 JAC85495.1 AK417962 BAN21177.1 APGK01051594 KB741194 ENN72989.1 KQ980304 KYN16739.1 GEDC01023622 JAS13676.1 GEZM01102783 GEZM01102782 JAV51795.1 LBMM01003520 KMQ93429.1 PZQS01000009 PVD24903.1 KQ435687 KOX81326.1 ADTU01028641 KZ288364 PBC26859.1 KQ981820 KYN35352.1 GL888644 EGI58669.1 KK854039 PTY10035.1 KQ414705 KOC63265.1 AMQM01000843 KB096742 ESO02315.1 GL447151 EFN86846.1 GL434909 EFN74326.1 KQ434783 KZC04976.1
KPJ15087.1 JTDY01000103 KOB78850.1 AGBW02009960 OWR49496.1 NWSH01000473 PCG76174.1 ODYU01002761 SOQ40658.1 GAIX01006120 JAA86440.1 KK852473 KDR23102.1 JXUM01122788 KQ566643 KXJ69952.1 NEVH01016952 PNF24875.1 GDAI01001527 JAI16076.1 GGFM01003328 MBW24079.1 GGFJ01010314 MBW59455.1 ADMH02001280 ETN63210.1 GGFJ01010313 MBW59454.1 GAMD01003176 JAA98414.1 CM000364 EDX12385.1 GGFK01007693 MBW41014.1 GBYB01003292 JAG73059.1 GAKP01015511 GAKP01015507 GAKP01015504 GAKP01015500 GAKP01015496 GAKP01015492 GAKP01015483 GAKP01015480 JAC43456.1 GFDG01001630 JAV17169.1 JRES01000657 KNC29480.1 UFQS01001990 UFQT01001990 SSX12886.1 SSX32328.1 APCN01000197 GAPW01005346 JAC08252.1 ATLV01011185 KE524650 KFB35904.1 CH480892 EDW55433.1 CM000160 EDW95978.1 AAAB01008807 EDO64594.1 CH954181 EDV48534.1 CH477478 EAT40260.1 AXCN02000387 CCAG010023983 CH940656 EDW58584.1 GFDL01002375 JAV32670.1 GAMC01000566 JAC05990.1 ABLF02017094 AF170829 AE014297 AY047546 KX531571 AAD49739.1 AAF55625.1 AAK77278.1 ANY27381.1 AXCM01000608 CH902617 EDV43647.1 CH933806 EDW16228.1 JXJN01024853 AJVK01035127 GALX01002889 JAB65577.1 CH916377 EDV90767.1 CH964272 EDW83640.1 CM000070 EAL26915.1 GFDG01003674 JAV15125.1 CH479186 EDW39124.1 GDRN01045575 JAI66840.1 GANO01004185 JAB55686.1 GBHO01033108 GBRD01014320 GDHC01012365 JAG10496.1 JAG51506.1 JAQ06264.1 AJWK01030327 GFDF01002131 JAV11953.1 CVRI01000043 CRK96206.1 PYGN01000545 PSN44502.1 KQ982254 KYQ58586.1 ACPB03022603 GBGD01003394 JAC85495.1 AK417962 BAN21177.1 APGK01051594 KB741194 ENN72989.1 KQ980304 KYN16739.1 GEDC01023622 JAS13676.1 GEZM01102783 GEZM01102782 JAV51795.1 LBMM01003520 KMQ93429.1 PZQS01000009 PVD24903.1 KQ435687 KOX81326.1 ADTU01028641 KZ288364 PBC26859.1 KQ981820 KYN35352.1 GL888644 EGI58669.1 KK854039 PTY10035.1 KQ414705 KOC63265.1 AMQM01000843 KB096742 ESO02315.1 GL447151 EFN86846.1 GL434909 EFN74326.1 KQ434783 KZC04976.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000007151
UP000218220
+ More
UP000027135 UP000069940 UP000249989 UP000235965 UP000000673 UP000075903 UP000000304 UP000075880 UP000075881 UP000037069 UP000075840 UP000030765 UP000001292 UP000095301 UP000002282 UP000075885 UP000075902 UP000095300 UP000076407 UP000075882 UP000007062 UP000076408 UP000008711 UP000008820 UP000075886 UP000092444 UP000008792 UP000069272 UP000007819 UP000000803 UP000075883 UP000007801 UP000078200 UP000009192 UP000075920 UP000192221 UP000092460 UP000092462 UP000091820 UP000001070 UP000075884 UP000007798 UP000001819 UP000008744 UP000075900 UP000092461 UP000092443 UP000183832 UP000245037 UP000075809 UP000192223 UP000015103 UP000019118 UP000079169 UP000078492 UP000036403 UP000245119 UP000053105 UP000005205 UP000242457 UP000078541 UP000007755 UP000053825 UP000015101 UP000008237 UP000000311 UP000076502
UP000027135 UP000069940 UP000249989 UP000235965 UP000000673 UP000075903 UP000000304 UP000075880 UP000075881 UP000037069 UP000075840 UP000030765 UP000001292 UP000095301 UP000002282 UP000075885 UP000075902 UP000095300 UP000076407 UP000075882 UP000007062 UP000076408 UP000008711 UP000008820 UP000075886 UP000092444 UP000008792 UP000069272 UP000007819 UP000000803 UP000075883 UP000007801 UP000078200 UP000009192 UP000075920 UP000192221 UP000092460 UP000092462 UP000091820 UP000001070 UP000075884 UP000007798 UP000001819 UP000008744 UP000075900 UP000092461 UP000092443 UP000183832 UP000245037 UP000075809 UP000192223 UP000015103 UP000019118 UP000079169 UP000078492 UP000036403 UP000245119 UP000053105 UP000005205 UP000242457 UP000078541 UP000007755 UP000053825 UP000015101 UP000008237 UP000000311 UP000076502
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JBG4
I4DK86
A0A194RBW0
A0A0L7LTQ9
A0A212F6X2
A0A2A4JX06
+ More
A0A2H1VIK0 S4PGP5 A0A067RH50 A0A182HAJ3 A0A2J7Q8H0 A0A0K8TPL1 A0A2M3Z6C9 A0A2M4C2E0 W5JJY1 A0A2M4C2D9 T1DFD6 A0A182VAH5 B4QTW2 A0A2M4AJS5 A0A0C9QRN3 A0A034VJH8 A0A182IK98 A0A182KIG7 A0A1L8EF04 A0A0L0CB83 A0A336L4P8 A0A182I347 A0A023EFY4 A0A084VD60 B4ILV7 A0A1I8MQN3 B4PMQ8 A0A182PQL2 A0A182TDQ3 A0A1I8P0Z4 A0A1Y9J0G2 A0A182KR52 A7URF1 A0A182XYQ0 B3P382 Q0IEU1 A0A182Q0I3 A0A1A9YUG3 B4MBW3 A0A182FDE3 A0A1Q3FYR9 W8CE99 J9K6Q7 Q9V3L7 A0A182MS06 B3LVB5 A0A1A9UJX9 B4K5Z1 A0A1Y9IWE2 A0A1W4UAV7 A0A1B0C382 A0A1B0DJQ5 A0A1A9W6P5 V5GV76 B4JYK5 A0A182NI19 B4NHZ0 Q29BT2 A0A1L8E937 B4GPP1 A0A182R7B6 A0A0N7ZDC2 U5EES5 A0A0A9WSU8 A0A1B0CV98 A0A1A9XN11 A0A1L8DZQ3 A0A1J1IB52 A0A2P8YJT3 A0A151XDY3 A0A1W4WQC4 T1I5A7 A0A069DNU3 R4WTB3 N6SYZ1 A0A1S3DAV6 A0A195DW01 A0A1B6CJL6 A0A1Y1JWN2 A0A0J7KT58 A0A2T7NUR1 A0A0M9ACY3 A0A158NWV3 A0A2A3E6R9 A0A195F5A2 F4X4F9 A0A2R7VVS7 A0A0L7QXD1 T1ERM6 E2BBS0 E1ZWY6 A0A154NZ72
A0A2H1VIK0 S4PGP5 A0A067RH50 A0A182HAJ3 A0A2J7Q8H0 A0A0K8TPL1 A0A2M3Z6C9 A0A2M4C2E0 W5JJY1 A0A2M4C2D9 T1DFD6 A0A182VAH5 B4QTW2 A0A2M4AJS5 A0A0C9QRN3 A0A034VJH8 A0A182IK98 A0A182KIG7 A0A1L8EF04 A0A0L0CB83 A0A336L4P8 A0A182I347 A0A023EFY4 A0A084VD60 B4ILV7 A0A1I8MQN3 B4PMQ8 A0A182PQL2 A0A182TDQ3 A0A1I8P0Z4 A0A1Y9J0G2 A0A182KR52 A7URF1 A0A182XYQ0 B3P382 Q0IEU1 A0A182Q0I3 A0A1A9YUG3 B4MBW3 A0A182FDE3 A0A1Q3FYR9 W8CE99 J9K6Q7 Q9V3L7 A0A182MS06 B3LVB5 A0A1A9UJX9 B4K5Z1 A0A1Y9IWE2 A0A1W4UAV7 A0A1B0C382 A0A1B0DJQ5 A0A1A9W6P5 V5GV76 B4JYK5 A0A182NI19 B4NHZ0 Q29BT2 A0A1L8E937 B4GPP1 A0A182R7B6 A0A0N7ZDC2 U5EES5 A0A0A9WSU8 A0A1B0CV98 A0A1A9XN11 A0A1L8DZQ3 A0A1J1IB52 A0A2P8YJT3 A0A151XDY3 A0A1W4WQC4 T1I5A7 A0A069DNU3 R4WTB3 N6SYZ1 A0A1S3DAV6 A0A195DW01 A0A1B6CJL6 A0A1Y1JWN2 A0A0J7KT58 A0A2T7NUR1 A0A0M9ACY3 A0A158NWV3 A0A2A3E6R9 A0A195F5A2 F4X4F9 A0A2R7VVS7 A0A0L7QXD1 T1ERM6 E2BBS0 E1ZWY6 A0A154NZ72
PDB
6G72
E-value=1.8452e-10,
Score=152
Ontologies
PATHWAY
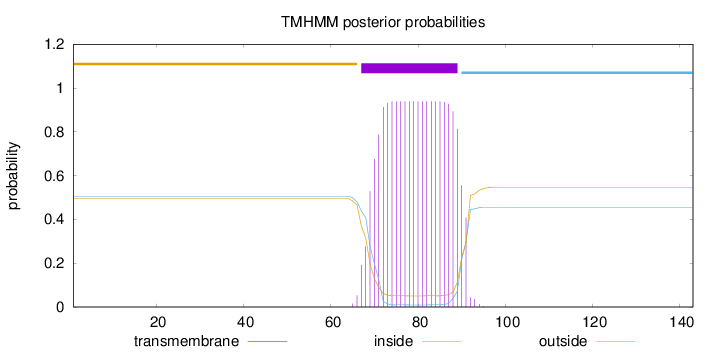
Topology
Length:
143
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.28008
Exp number, first 60 AAs:
0
Total prob of N-in:
0.50416
outside
1 - 66
TMhelix
67 - 89
inside
90 - 143
Population Genetic Test Statistics
Pi
4.076146
Theta
11.695287
Tajima's D
-1.931811
CLR
245.284992
CSRT
0.0143992800359982
Interpretation
Possibly Positive selection
