Gene
KWMTBOMO05699 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006828
Annotation
PREDICTED:_tRNA-splicing_ligase_RtcB_homolog_[Amyelois_transitella]
Full name
tRNA-splicing ligase RtcB homolog
+ More
tRNA-splicing ligase RtcB homolog 1
tRNA-splicing ligase RtcB homolog 1
Location in the cell
Cytoplasmic Reliability : 2.375 Mitochondrial Reliability : 1.988
Sequence
CDS
ATGGTTGTAAGGCAGTATAATGAAGAAATTAAATATTTGGAAAAAATTAATCCATATTGTTGGAGAATTAAAAAAGGCTTCCAACCAAATATGAATGTTGAAGGAGTGTTTTACGTTAACAGTACACTTGAGAAACTAATGTTGGAGGAGCTTAGGAATTGTTGTAGACCTGGTATGTCAGGTGGATTCCTTCCTGGTGTAAAACAGATAGCTAATGTTGCAGCTTTACCGGGAATTGTAGGCCGGTCAGTGGGATTGCCTGATGTTCATTCTGGATATGGGTTCGCTATAGGAAATATGGCAGCTTTTGATATGGATGATCCCAAATCTATAGTATCTCCAGGAGGCGTAGGATTCGATATAAACTGTGGTGTTCGTTTATTGAGAACAAATTTACATGAGAAGGATGTTCTACCTATTAAGGAACAATTAGCTCAGAGCTTGTTTGACCATATACCTGTTGGAGTTGGCTCCAAGGGTATTATTCCAATGAATGCACGTGATTTGGAAGAAGCATTAGAAATGGGTATGGACTGGTCTCTAAGGGAAGGTTATGTTTGGGCCGAAGATAAGGAACACTGTGAAGAATATGGAAGAATGTTGAATGCTGATCCATCAAAAGTCAGTCTCAGAGCAAAGAAAAGAGGTCTACCTCAACTGGGCACACTAGGAGCTGGAAATCATTATGCAGAGATTCAAGTTGTTGATGAAATATTTGATAAATATGCTGCTGGAAAAATGGGTATTGAAAGTATTGGTCAAGTTTGTGTGATGATTCACTCTGGAAGTCGTGGTTTTGGGCATCAAGTAGCAACTGATGCTTTAGTGCAAATGGAGAAAGCTATGAAGAGAGATAACATAGAAACTAATGATAGACAACTTGCATGTGCCAGGATAAAATCTGTTGAAGGTCAAGATTATCTTAAAGCTATGTCTGCTGCTGCCAATTTTGCTTGGGTGAACCGTAGCTCAATGACATTCTTGACCCGTCAAGCTTTTGCGAAGCAATTTAAAATGGCACCTGATGATCTTGACATGCATGTCATATATGATGTGTCTCATAATATTGCCAAGGTTGAAGAACATGTAGTAGATGGGAAATTGAAAACTCTACTAGTACATAGGAAGGGCTCAACTAGAGCATTCCCACCAAATCATCCATTAATACCTGTGGATTATCAACTAACTGGTCAGCCTGTACTTATTGGTGGTACAATGGGAACCTGCAGTTATGTACTAACTGGTACTCATCAAGGAATGACAGAAACGTTTGGATCCACATGCCATGGTGCTGGTCGAGCTCTGTCTAGAGCCAAGTCTAGAAGAAACATAGACTACAAAGAAGTTCTAAACAAACTTGAAACTATGGGTATTTCCATTAGAGTTGCATCTCCAAAACTAGTAATGGAAGAGGCACCAGAGTCATACAAGAATGTAACAGATGTCGTAGATACATGTCATGCTGCTGGTATTAGCAAAAAAACTGTTAAATTAAGACCAATTGCAGTTATAAAAGGATGA
Protein
MVVRQYNEEIKYLEKINPYCWRIKKGFQPNMNVEGVFYVNSTLEKLMLEELRNCCRPGMSGGFLPGVKQIANVAALPGIVGRSVGLPDVHSGYGFAIGNMAAFDMDDPKSIVSPGGVGFDINCGVRLLRTNLHEKDVLPIKEQLAQSLFDHIPVGVGSKGIIPMNARDLEEALEMGMDWSLREGYVWAEDKEHCEEYGRMLNADPSKVSLRAKKRGLPQLGTLGAGNHYAEIQVVDEIFDKYAAGKMGIESIGQVCVMIHSGSRGFGHQVATDALVQMEKAMKRDNIETNDRQLACARIKSVEGQDYLKAMSAAANFAWVNRSSMTFLTRQAFAKQFKMAPDDLDMHVIYDVSHNIAKVEEHVVDGKLKTLLVHRKGSTRAFPPNHPLIPVDYQLTGQPVLIGGTMGTCSYVLTGTHQGMTETFGSTCHGAGRALSRAKSRRNIDYKEVLNKLETMGISIRVASPKLVMEEAPESYKNVTDVVDTCHAAGISKKTVKLRPIAVIKG
Summary
Description
Catalytic subunit of the tRNA-splicing ligase complex that acts by directly joining spliced tRNA halves to mature-sized tRNAs by incorporating the precursor-derived splice junction phosphate into the mature tRNA as a canonical 3',5'-phosphodiester. May act as a RNA ligase with broad substrate specificity, and may function toward other RNAs.
Catalytic subunit of the tRNA-splicing ligase complex that acts by directly joining spliced tRNA halves to mature-sized tRNAs by incorporating the precursor-derived splice junction phosphate into the mature tRNA as a canonical 3',5'-phosphodiester. May act as an RNA ligase with broad substrate specificity, and may function toward other RNAs.
Catalytic subunit of the tRNA-splicing ligase complex that acts by directly joining spliced tRNA halves to mature-sized tRNAs by incorporating the precursor-derived splice junction phosphate into the mature tRNA as a canonical 3',5'-phosphodiester. May act as an RNA ligase with broad substrate specificity, and may function toward other RNAs.
Catalytic Activity
ATP + (ribonucleotide)(n)-3'-hydroxyl + 5'-phospho-(ribonucleotide)(m) = (ribonucleotide)(n+m) + AMP + diphosphate.
Cofactor
Mn(2+)
Mg(2+)
Mg(2+)
Subunit
Catalytic component of the tRNA-splicing ligase complex.
Miscellaneous
Ligation probably proceeds through 3 nucleotidyl transfer steps, with 2',3'-cyclic phosphate termini being hydrolyzed to 3'-P termini in a step that precedes 3'-P activation with GMP. In the first nucleotidyl transfer step, RTCB reacts with GTP to form a covalent RTCB-histidine-GMP intermediate with release of PPi; in the second step, the GMP moiety is transferred to the RNA 3'-P; in the third step, the 5'-OH from the opposite RNA strand attacks the activated 3'-P to form a 3',5'-phosphodiester bond and release GMP (By similarity).
Similarity
Belongs to the RtcB family.
Belongs to the PP2C family.
Belongs to the PP2C family.
Keywords
ATP-binding
Complete proteome
Ligase
Manganese
Metal-binding
Nucleotide-binding
Reference proteome
tRNA processing
Feature
chain tRNA-splicing ligase RtcB homolog
Uniprot
H9JBD3
A0A2A4JXE7
S4P2I1
A0A067R6Q5
A0A2J7PJX5
D6X2M2
+ More
A0A194RB85 A0A212F6Z6 A0A194PDY6 A0A1W4WJG3 A0A1B6MSQ8 J3JXR6 E0W052 A0A0A9YMD2 A0A1Y1JYW8 A0A1B0G046 A0A232EPJ7 A0A026WKW4 K7IRR0 E2BMV5 E2AA83 A0A2H1VIK8 A0A2A3ERE5 A0A1I8PP30 A0A088AVQ8 A0A0P4WBJ9 A0A1A9X854 A0A1A9V1A8 A0A0V0GAS7 A0A069DUS0 A0A158NRH5 T1I6I8 A0A023F4W9 A0A1B0A883 F4WBN8 A0A151IK44 A0A2H8TWY2 A0A0L7LUE5 A0A0L0BW38 V5HPV6 A0A2S2R6J2 J9JML1 A0A1W4VFA5 B4PAY3 A0A0M5J6I3 A0A3B0KGL5 B3MK04 B4M963 A0A1L8DAT5 A0A1I8MLA1 Q29K48 B4GWW3 B3NM58 B4MTG4 A0A1B0D6J7 A0A0J9R4J8 A0A154PN65 B4I5U9 A0A2M4AMA7 W5JBW7 A0A084W855 B4KK68 Q9VIW7 A0A1E1X3E6 B4Q9X8 B0WCT9 A0A131XP60 A0A224Z4C6 A0A182MQS9 B4JP85 A0A182W707 A0A182SJ88 W8CA79 A0A182RCS7 A0A182YQX8 A0A0A1XE59 A0A182QK70 A0A182V991 A0A182UFB1 A0A182L1D4 A0A182WU02 A0A182HUL8 Q7Q412 A0A2R5LIC6 A0A1Q3FA44 A0A182NUG2 A0A131Z1G4 A0A182JPP6 A0A023GCS6 A0A182PPK9 A0A182F4X4 A0A0P5EUL4 A0A0K8TW85 A0A034W5W1 A0A023FKH8 T1IMQ9 A0A1D2MN93 A0A0P5MQ97 E9HNI6 A0A336LXW7
A0A194RB85 A0A212F6Z6 A0A194PDY6 A0A1W4WJG3 A0A1B6MSQ8 J3JXR6 E0W052 A0A0A9YMD2 A0A1Y1JYW8 A0A1B0G046 A0A232EPJ7 A0A026WKW4 K7IRR0 E2BMV5 E2AA83 A0A2H1VIK8 A0A2A3ERE5 A0A1I8PP30 A0A088AVQ8 A0A0P4WBJ9 A0A1A9X854 A0A1A9V1A8 A0A0V0GAS7 A0A069DUS0 A0A158NRH5 T1I6I8 A0A023F4W9 A0A1B0A883 F4WBN8 A0A151IK44 A0A2H8TWY2 A0A0L7LUE5 A0A0L0BW38 V5HPV6 A0A2S2R6J2 J9JML1 A0A1W4VFA5 B4PAY3 A0A0M5J6I3 A0A3B0KGL5 B3MK04 B4M963 A0A1L8DAT5 A0A1I8MLA1 Q29K48 B4GWW3 B3NM58 B4MTG4 A0A1B0D6J7 A0A0J9R4J8 A0A154PN65 B4I5U9 A0A2M4AMA7 W5JBW7 A0A084W855 B4KK68 Q9VIW7 A0A1E1X3E6 B4Q9X8 B0WCT9 A0A131XP60 A0A224Z4C6 A0A182MQS9 B4JP85 A0A182W707 A0A182SJ88 W8CA79 A0A182RCS7 A0A182YQX8 A0A0A1XE59 A0A182QK70 A0A182V991 A0A182UFB1 A0A182L1D4 A0A182WU02 A0A182HUL8 Q7Q412 A0A2R5LIC6 A0A1Q3FA44 A0A182NUG2 A0A131Z1G4 A0A182JPP6 A0A023GCS6 A0A182PPK9 A0A182F4X4 A0A0P5EUL4 A0A0K8TW85 A0A034W5W1 A0A023FKH8 T1IMQ9 A0A1D2MN93 A0A0P5MQ97 E9HNI6 A0A336LXW7
EC Number
6.5.1.3
Pubmed
19121390
23622113
24845553
18362917
19820115
26354079
+ More
22118469 22516182 20566863 25401762 26823975 28004739 28648823 24508170 30249741 20075255 20798317 26334808 21347285 25474469 21719571 26227816 26108605 25765539 17994087 17550304 25315136 15632085 22936249 20920257 23761445 24438588 18057021 10731132 12537572 12537569 28503490 28049606 28797301 24495485 25244985 25830018 20966253 12364791 26830274 25348373 27289101 21292972
22118469 22516182 20566863 25401762 26823975 28004739 28648823 24508170 30249741 20075255 20798317 26334808 21347285 25474469 21719571 26227816 26108605 25765539 17994087 17550304 25315136 15632085 22936249 20920257 23761445 24438588 18057021 10731132 12537572 12537569 28503490 28049606 28797301 24495485 25244985 25830018 20966253 12364791 26830274 25348373 27289101 21292972
EMBL
BABH01030873
NWSH01000473
PCG76173.1
GAIX01012005
JAA80555.1
KK852704
+ More
KDR18043.1 NEVH01024944 PNF16636.1 KQ971372 EFA09424.1 KQ460398 KPJ15088.1 AGBW02009960 OWR49497.1 KQ459606 KPI91472.1 GEBQ01001052 JAT38925.1 BT128037 AEE62998.1 AAZO01006625 DS235855 EEB19008.1 GBHO01009402 GDHC01004341 JAG34202.1 JAQ14288.1 GEZM01102990 JAV51637.1 CCAG010002365 NNAY01002948 OXU20236.1 KK107167 QOIP01000010 EZA56311.1 RLU18191.1 GL449347 EFN82990.1 GL438030 EFN69663.1 ODYU01002761 SOQ40657.1 KZ288198 PBC33699.1 GDRN01071189 JAI63738.1 GECL01000841 JAP05283.1 GBGD01001204 JAC87685.1 ADTU01024032 ACPB03011590 GBBI01002181 JAC16531.1 GL888066 EGI68316.1 KQ977255 KYN04645.1 GFXV01007000 MBW18805.1 JTDY01000103 KOB78846.1 JRES01001249 KNC24242.1 GANP01006686 JAB77782.1 GGMS01016361 MBY85564.1 ABLF02037893 CM000158 EDW90412.1 CP012523 ALC39974.1 OUUW01000010 SPP85489.1 CH902620 EDV32459.1 CH940654 EDW57739.1 GFDF01010515 JAV03569.1 CH379061 EAL33330.1 CH479195 EDW27290.1 CH954179 EDV54658.1 CH963852 EDW75403.1 AJVK01003665 CM002910 KMY91008.1 KQ434998 KZC13306.1 CH480822 EDW55755.1 GGFK01008556 MBW41877.1 ADMH02001944 ETN60385.1 ATLV01021368 KE525317 KFB46399.1 CH933807 EDW11586.1 KRG02784.1 AE014134 AY119213 GFAC01005426 JAT93762.1 CM000361 EDX05508.1 DS231890 GEFH01000671 JAP67910.1 GFPF01012879 MAA24025.1 AXCM01008504 CH916372 EDV98715.1 GAMC01001519 JAC05037.1 GBXI01005062 JAD09230.1 AXCN02000761 APCN01002465 AAAB01008964 GGLE01005148 MBY09274.1 GFDL01010590 JAV24455.1 GEDV01004576 JAP83981.1 GBBM01003739 JAC31679.1 GDIP01143132 LRGB01002860 JAJ80270.1 KZS05822.1 GDHF01033756 JAI18558.1 GAKP01007976 JAC50976.1 GBBK01002345 JAC22137.1 JH431093 LJIJ01000802 ODM94463.1 GDIQ01152934 JAK98791.1 GL732696 EFX66707.1 UFQS01000215 UFQT01000215 SSX01451.1 SSX21831.1
KDR18043.1 NEVH01024944 PNF16636.1 KQ971372 EFA09424.1 KQ460398 KPJ15088.1 AGBW02009960 OWR49497.1 KQ459606 KPI91472.1 GEBQ01001052 JAT38925.1 BT128037 AEE62998.1 AAZO01006625 DS235855 EEB19008.1 GBHO01009402 GDHC01004341 JAG34202.1 JAQ14288.1 GEZM01102990 JAV51637.1 CCAG010002365 NNAY01002948 OXU20236.1 KK107167 QOIP01000010 EZA56311.1 RLU18191.1 GL449347 EFN82990.1 GL438030 EFN69663.1 ODYU01002761 SOQ40657.1 KZ288198 PBC33699.1 GDRN01071189 JAI63738.1 GECL01000841 JAP05283.1 GBGD01001204 JAC87685.1 ADTU01024032 ACPB03011590 GBBI01002181 JAC16531.1 GL888066 EGI68316.1 KQ977255 KYN04645.1 GFXV01007000 MBW18805.1 JTDY01000103 KOB78846.1 JRES01001249 KNC24242.1 GANP01006686 JAB77782.1 GGMS01016361 MBY85564.1 ABLF02037893 CM000158 EDW90412.1 CP012523 ALC39974.1 OUUW01000010 SPP85489.1 CH902620 EDV32459.1 CH940654 EDW57739.1 GFDF01010515 JAV03569.1 CH379061 EAL33330.1 CH479195 EDW27290.1 CH954179 EDV54658.1 CH963852 EDW75403.1 AJVK01003665 CM002910 KMY91008.1 KQ434998 KZC13306.1 CH480822 EDW55755.1 GGFK01008556 MBW41877.1 ADMH02001944 ETN60385.1 ATLV01021368 KE525317 KFB46399.1 CH933807 EDW11586.1 KRG02784.1 AE014134 AY119213 GFAC01005426 JAT93762.1 CM000361 EDX05508.1 DS231890 GEFH01000671 JAP67910.1 GFPF01012879 MAA24025.1 AXCM01008504 CH916372 EDV98715.1 GAMC01001519 JAC05037.1 GBXI01005062 JAD09230.1 AXCN02000761 APCN01002465 AAAB01008964 GGLE01005148 MBY09274.1 GFDL01010590 JAV24455.1 GEDV01004576 JAP83981.1 GBBM01003739 JAC31679.1 GDIP01143132 LRGB01002860 JAJ80270.1 KZS05822.1 GDHF01033756 JAI18558.1 GAKP01007976 JAC50976.1 GBBK01002345 JAC22137.1 JH431093 LJIJ01000802 ODM94463.1 GDIQ01152934 JAK98791.1 GL732696 EFX66707.1 UFQS01000215 UFQT01000215 SSX01451.1 SSX21831.1
Proteomes
UP000005204
UP000218220
UP000027135
UP000235965
UP000007266
UP000053240
+ More
UP000007151 UP000053268 UP000192223 UP000009046 UP000092444 UP000215335 UP000053097 UP000279307 UP000002358 UP000008237 UP000000311 UP000242457 UP000095300 UP000005203 UP000092443 UP000078200 UP000005205 UP000015103 UP000092445 UP000007755 UP000078542 UP000037510 UP000037069 UP000007819 UP000192221 UP000002282 UP000092553 UP000268350 UP000007801 UP000008792 UP000095301 UP000001819 UP000008744 UP000008711 UP000007798 UP000092462 UP000076502 UP000001292 UP000000673 UP000030765 UP000009192 UP000000803 UP000000304 UP000002320 UP000075883 UP000001070 UP000075920 UP000075901 UP000075900 UP000076408 UP000075886 UP000075903 UP000075902 UP000075882 UP000076407 UP000075840 UP000007062 UP000075884 UP000075881 UP000075885 UP000069272 UP000076858 UP000094527 UP000000305
UP000007151 UP000053268 UP000192223 UP000009046 UP000092444 UP000215335 UP000053097 UP000279307 UP000002358 UP000008237 UP000000311 UP000242457 UP000095300 UP000005203 UP000092443 UP000078200 UP000005205 UP000015103 UP000092445 UP000007755 UP000078542 UP000037510 UP000037069 UP000007819 UP000192221 UP000002282 UP000092553 UP000268350 UP000007801 UP000008792 UP000095301 UP000001819 UP000008744 UP000008711 UP000007798 UP000092462 UP000076502 UP000001292 UP000000673 UP000030765 UP000009192 UP000000803 UP000000304 UP000002320 UP000075883 UP000001070 UP000075920 UP000075901 UP000075900 UP000076408 UP000075886 UP000075903 UP000075902 UP000075882 UP000076407 UP000075840 UP000007062 UP000075884 UP000075881 UP000075885 UP000069272 UP000076858 UP000094527 UP000000305
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JBD3
A0A2A4JXE7
S4P2I1
A0A067R6Q5
A0A2J7PJX5
D6X2M2
+ More
A0A194RB85 A0A212F6Z6 A0A194PDY6 A0A1W4WJG3 A0A1B6MSQ8 J3JXR6 E0W052 A0A0A9YMD2 A0A1Y1JYW8 A0A1B0G046 A0A232EPJ7 A0A026WKW4 K7IRR0 E2BMV5 E2AA83 A0A2H1VIK8 A0A2A3ERE5 A0A1I8PP30 A0A088AVQ8 A0A0P4WBJ9 A0A1A9X854 A0A1A9V1A8 A0A0V0GAS7 A0A069DUS0 A0A158NRH5 T1I6I8 A0A023F4W9 A0A1B0A883 F4WBN8 A0A151IK44 A0A2H8TWY2 A0A0L7LUE5 A0A0L0BW38 V5HPV6 A0A2S2R6J2 J9JML1 A0A1W4VFA5 B4PAY3 A0A0M5J6I3 A0A3B0KGL5 B3MK04 B4M963 A0A1L8DAT5 A0A1I8MLA1 Q29K48 B4GWW3 B3NM58 B4MTG4 A0A1B0D6J7 A0A0J9R4J8 A0A154PN65 B4I5U9 A0A2M4AMA7 W5JBW7 A0A084W855 B4KK68 Q9VIW7 A0A1E1X3E6 B4Q9X8 B0WCT9 A0A131XP60 A0A224Z4C6 A0A182MQS9 B4JP85 A0A182W707 A0A182SJ88 W8CA79 A0A182RCS7 A0A182YQX8 A0A0A1XE59 A0A182QK70 A0A182V991 A0A182UFB1 A0A182L1D4 A0A182WU02 A0A182HUL8 Q7Q412 A0A2R5LIC6 A0A1Q3FA44 A0A182NUG2 A0A131Z1G4 A0A182JPP6 A0A023GCS6 A0A182PPK9 A0A182F4X4 A0A0P5EUL4 A0A0K8TW85 A0A034W5W1 A0A023FKH8 T1IMQ9 A0A1D2MN93 A0A0P5MQ97 E9HNI6 A0A336LXW7
A0A194RB85 A0A212F6Z6 A0A194PDY6 A0A1W4WJG3 A0A1B6MSQ8 J3JXR6 E0W052 A0A0A9YMD2 A0A1Y1JYW8 A0A1B0G046 A0A232EPJ7 A0A026WKW4 K7IRR0 E2BMV5 E2AA83 A0A2H1VIK8 A0A2A3ERE5 A0A1I8PP30 A0A088AVQ8 A0A0P4WBJ9 A0A1A9X854 A0A1A9V1A8 A0A0V0GAS7 A0A069DUS0 A0A158NRH5 T1I6I8 A0A023F4W9 A0A1B0A883 F4WBN8 A0A151IK44 A0A2H8TWY2 A0A0L7LUE5 A0A0L0BW38 V5HPV6 A0A2S2R6J2 J9JML1 A0A1W4VFA5 B4PAY3 A0A0M5J6I3 A0A3B0KGL5 B3MK04 B4M963 A0A1L8DAT5 A0A1I8MLA1 Q29K48 B4GWW3 B3NM58 B4MTG4 A0A1B0D6J7 A0A0J9R4J8 A0A154PN65 B4I5U9 A0A2M4AMA7 W5JBW7 A0A084W855 B4KK68 Q9VIW7 A0A1E1X3E6 B4Q9X8 B0WCT9 A0A131XP60 A0A224Z4C6 A0A182MQS9 B4JP85 A0A182W707 A0A182SJ88 W8CA79 A0A182RCS7 A0A182YQX8 A0A0A1XE59 A0A182QK70 A0A182V991 A0A182UFB1 A0A182L1D4 A0A182WU02 A0A182HUL8 Q7Q412 A0A2R5LIC6 A0A1Q3FA44 A0A182NUG2 A0A131Z1G4 A0A182JPP6 A0A023GCS6 A0A182PPK9 A0A182F4X4 A0A0P5EUL4 A0A0K8TW85 A0A034W5W1 A0A023FKH8 T1IMQ9 A0A1D2MN93 A0A0P5MQ97 E9HNI6 A0A336LXW7
PDB
4IT0
E-value=6.68519e-128,
Score=1172
Ontologies
GO
PANTHER
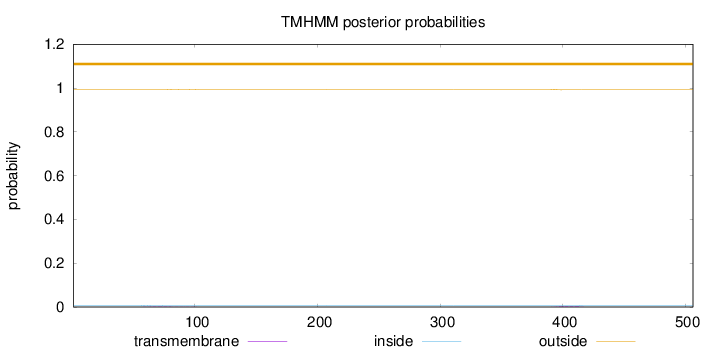
Topology
Length:
506
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0992700000000001
Exp number, first 60 AAs:
0.00235
Total prob of N-in:
0.00756
outside
1 - 506
Population Genetic Test Statistics
Pi
228.5173
Theta
198.211153
Tajima's D
0.590671
CLR
0.370892
CSRT
0.538773061346933
Interpretation
Uncertain
