Gene
KWMTBOMO05697
Pre Gene Modal
BGIBMGA006857
Annotation
AF032873_1_inebriated_protein_[Manduca_sexta]
Full name
Transporter
+ More
Sodium- and chloride-dependent GABA transporter ine
Sodium- and chloride-dependent GABA transporter ine
Alternative Name
Protein inebriated
Protein receptor oscillation A
Protein receptor oscillation A
Location in the cell
PlasmaMembrane Reliability : 4.969
Sequence
CDS
ATGAACAAAGCCGAATCTTCGACGGAAACCGCCGCGCCCAGCGTGGCGATCCACGTGGAACAGTATGACGACTGTCAGGACGTCGAAAGCTCAAAGCTTCTCAACCTGCAAGAACCGGCAGCTACAATTAATACCTCTGCACCAATGAGAAAAGTTAAGAGCTTTAGTGACACCCAAAAAATTCGTGATGTTGCCGCTGCTTCCGGAGCTGCTTCTGCAAGAAGTTTACGACCATATGAACAAGTGATTCCTTACCCTGAAGGATCCGAAAACGGAACTAACCAATATGGAGCGCCATCGGTTCGATCCCTCGCTTCAATCGGCATGGGCTGCACCGATGGTCGCAAAATGGTTATTAGACGAGTACCTACTTCGCCCACAGAATTGTTCCATCTAGTGCGTCCTCCTACACCCCCCGATGAAGATTCGGCATCCCACGAGAGTGACTGTGAAGAAGAAGAAGAAGATGCCGCAGTACATTTGAAACCCCGAAGGCCCTTTTGGGCTAACAAGATTCAGTTCGTGTTGGCATGCGTCGGATACTCCGTTGGCCTCGGCAATGTTTGGCGCTTTCCTTATCTCTGTTACAAAAGTGGAGGAGGTGCATTTTTGATACCTTACTTCATAATATTGTTAGTTTGCGGCGTCCCGATGCTCTTTATGGAACTGGCTATAGGGCAATACACAGCTCACGGACCTATAGGAGCTTTGTCGCAGATATGTCCACTTTTTAAAGGTGCAGGTTTAGCGAGTGTGGTGATCTCATTTTTAATGTCCACATACTATGCTGTGATAATCGCGTGGGCAATTTATTATTTCTTCACGTCTTTCAAAACGGAAGTACCATGGGCCAGTTGTTCAAATCGATGGAACACGGATATGTGCTGGGTGCCAAACCATAACTACACAAAACCGAATGGATCTCAAACACCCACCGAACAATTCTTCGAGAAGAAGGTATTGAGTATGAGTGCCGGTATAGAATTTCCTGGTGGCATGCGGTGGGAATTGGCAGCTTGCTTGGTCTGTGCTTGGGTGTTAGTTTATTTCGCCCTGTGGAAAAGTATCAAGTCATCCGCCAAAGTTCGTTATATCACCACGACACTGCCTTTTCTGCTAATTTTAGTATTCTTAGGTAGATCTCTAACACTCGATGGGGCGGACAGAGGCTTGAGATTTTTCTTCAAACCAGAATGGGAACTTCTAAAACAATCCAGACCATGGGTTAACGCCGCTTCACAAATTTTCAATTCCATCGGCATAGCGTTCGGATCTATGATCATGTTCGCTTCATATAACAGATTCGACAACAATTTTCTACACGATACTGTGGCTGTAACATTGGTCAATGCAATAACCAGTCTCATAGTCGGAATATTCACCTTCGCTACTATTGGAAATATCGCATTTGAACAAAACACACCGGTCAAAGATGTTATCGCAGATAGTCCCGGACTTTTATTCGTGGTTTATCCACAAGCAATTGCAAAGATGCCAGCTTCGCAACTTTGGGCAGTTTTATTTTTCTTTATGTTCCTGTGCCTTGGCCTGAACAGTCAGTTTGCCATTGTGGAAGTTGTCGTTACTTCGATACAAGATGGTTTCCCAGATATGATCCGTAGGAAACTTGTCTATCATGAGCTCTTAGTATTATTGGTGTGCGCGGTGTCACTCGTATTCGGCCTACCACATATAATACATAGCGGCATATACATATTCCAATTAATGGACTACTATGCCGCTTCGCTCAGTATCACGTATCTTGCATTTTTCGAAGTTGTTGCCATAGCTTGGTTCTATGGAGTGGGGAGACTTTCTCGAAACATTAAACAGATGACAGGTCGACGTCCGTCAATTTATTTCCGATTTTGTTGGCTGATTGCTACACCTGCTCTATTAATAGCACTATGGGTGGCAAGTCTCGTCGATTATACTCCCCCCAGCTACCGGCAGTACCAATATCCAGCATGGGCACAAGCAATTGGTTGGATTATCGCCTCCCTTTCCTTATTATGCATTCCTGTGTACGCTGTAATAGTCGTATTTAGAGCACCCGGGGAAAATTTACTTGAGAAATTACGTTATTCCATTCGCCCGAGTTCTATTTGCGACTGTGGCGTGAATGGTTGTGATATATGCTGCTCGGAATCCGAAACTCCTGAAGATAAAACTGTTATAACTTAG
Protein
MNKAESSTETAAPSVAIHVEQYDDCQDVESSKLLNLQEPAATINTSAPMRKVKSFSDTQKIRDVAAASGAASARSLRPYEQVIPYPEGSENGTNQYGAPSVRSLASIGMGCTDGRKMVIRRVPTSPTELFHLVRPPTPPDEDSASHESDCEEEEEDAAVHLKPRRPFWANKIQFVLACVGYSVGLGNVWRFPYLCYKSGGGAFLIPYFIILLVCGVPMLFMELAIGQYTAHGPIGALSQICPLFKGAGLASVVISFLMSTYYAVIIAWAIYYFFTSFKTEVPWASCSNRWNTDMCWVPNHNYTKPNGSQTPTEQFFEKKVLSMSAGIEFPGGMRWELAACLVCAWVLVYFALWKSIKSSAKVRYITTTLPFLLILVFLGRSLTLDGADRGLRFFFKPEWELLKQSRPWVNAASQIFNSIGIAFGSMIMFASYNRFDNNFLHDTVAVTLVNAITSLIVGIFTFATIGNIAFEQNTPVKDVIADSPGLLFVVYPQAIAKMPASQLWAVLFFFMFLCLGLNSQFAIVEVVVTSIQDGFPDMIRRKLVYHELLVLLVCAVSLVFGLPHIIHSGIYIFQLMDYYAASLSITYLAFFEVVAIAWFYGVGRLSRNIKQMTGRRPSIYFRFCWLIATPALLIALWVASLVDYTPPSYRQYQYPAWAQAIGWIIASLSLLCIPVYAVIVVFRAPGENLLEKLRYSIRPSSICDCGVNGCDICCSESETPEDKTVIT
Summary
Description
Plays a role in neuronal membrane excitation, important for normal response properties of the photoreceptor. Able to control excitability from either neurons or glia cells. Ine negatively regulates neuronal sodium channels. Controls neurotransmitter-mediated signaling pathways associated with the structure of the larval peripheral nerve, ine and eag control perineurial glial growth through partially redundant pathways. Isoform A and isoform B are both functional, although isoform A functions with greater efficiency. Has a role in osmolyte transport within the Malpighian tubule and hindgut.
Similarity
Belongs to the sodium:neurotransmitter symporter (SNF) (TC 2.A.22) family.
Keywords
Alternative splicing
Complete proteome
Developmental protein
Differentiation
Glycoprotein
Membrane
Neurogenesis
Neurotransmitter transport
Reference proteome
Symport
Transmembrane
Transmembrane helix
Transport
Feature
chain Sodium- and chloride-dependent GABA transporter ine
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9JBG2
Q9U5A5
A0A2H1VK69
A0A194RBW5
A0A194PK53
A0A0L7LTT3
+ More
A0A2A4JWJ5 A0A212F710 A0A1L8DJZ1 A0A182WG85 A0A1S4GZP7 A0A182Y0E7 A0A2M4CIM3 A0A2M4CIC0 U5EUA5 A0A1Q3FUS3 A0A1Q3FQM0 A0A1Q3FQP0 B4LUW5 D6X2M1 A0A0A1X708 B4MTX2 T1PET9 A0A0L0C6X9 A0A1I8NQS7 A0A1I8MTT9 B4NYY7 B3MU16 A0A1W4WA05 K7J4G0 A0A0J9QVL8 A0A1I8NQS8 B4I3A5 B3N3T6 B4KLF2 A0A0K8UTA4 E1JHT2 Q9VR07 E2BGZ3 A0A1Y1L6M7 A0A3B0K6Z8 B4GQE8 Q29LA0 A0A182P1M5 B4JQR8 A0A026WPJ4 A0A182I7E3 E2A8H3 A0A182NUS7 A0A310SJ50 A0A084W1U8 A0A2A3EH09 A0A1A9VW31 A0A195BBM1 A0A1S4GZP4 A0A182LUR7 A0A195D5A3 A0A1A9XJP4 A0A088A4T6 A0A1B0C304 X2D4U8 A0A195DXM1 A0A182F1I2 A0A182QL93 A0A158NJT5 A0A182J770 W5JAR1 Q7YZC7 A0A182LMQ1 F4X6F1 A0A1Q3FQS7 A0A182UTC3 A0A1Q3FR06 Q7PWJ5 B0WB25 A0A195FMG1 A0A182G8W1 A0A1A9ZCS2 A0A1J1IWZ2 A0A0Q9WA98 E9IFV1 A0A232FDE8 A0A154P7S6 A0A336MFN3 A0A0R1DJ70 A0A0M4E6D1 A0A1B6CQI5 A0A0P9A7T6 A0A1W4VY60 A0A1I8M2G9 A0A151X6F1 A0A0J9TFF7 A0A0Q9X5I5 A0A0Q5W9A0 A0A0A1XCF1 A0A1B6LKH7 Q9VR07-2 C8VV47 A0A3B0JGD1 A0A0K8WJF4
A0A2A4JWJ5 A0A212F710 A0A1L8DJZ1 A0A182WG85 A0A1S4GZP7 A0A182Y0E7 A0A2M4CIM3 A0A2M4CIC0 U5EUA5 A0A1Q3FUS3 A0A1Q3FQM0 A0A1Q3FQP0 B4LUW5 D6X2M1 A0A0A1X708 B4MTX2 T1PET9 A0A0L0C6X9 A0A1I8NQS7 A0A1I8MTT9 B4NYY7 B3MU16 A0A1W4WA05 K7J4G0 A0A0J9QVL8 A0A1I8NQS8 B4I3A5 B3N3T6 B4KLF2 A0A0K8UTA4 E1JHT2 Q9VR07 E2BGZ3 A0A1Y1L6M7 A0A3B0K6Z8 B4GQE8 Q29LA0 A0A182P1M5 B4JQR8 A0A026WPJ4 A0A182I7E3 E2A8H3 A0A182NUS7 A0A310SJ50 A0A084W1U8 A0A2A3EH09 A0A1A9VW31 A0A195BBM1 A0A1S4GZP4 A0A182LUR7 A0A195D5A3 A0A1A9XJP4 A0A088A4T6 A0A1B0C304 X2D4U8 A0A195DXM1 A0A182F1I2 A0A182QL93 A0A158NJT5 A0A182J770 W5JAR1 Q7YZC7 A0A182LMQ1 F4X6F1 A0A1Q3FQS7 A0A182UTC3 A0A1Q3FR06 Q7PWJ5 B0WB25 A0A195FMG1 A0A182G8W1 A0A1A9ZCS2 A0A1J1IWZ2 A0A0Q9WA98 E9IFV1 A0A232FDE8 A0A154P7S6 A0A336MFN3 A0A0R1DJ70 A0A0M4E6D1 A0A1B6CQI5 A0A0P9A7T6 A0A1W4VY60 A0A1I8M2G9 A0A151X6F1 A0A0J9TFF7 A0A0Q9X5I5 A0A0Q5W9A0 A0A0A1XCF1 A0A1B6LKH7 Q9VR07-2 C8VV47 A0A3B0JGD1 A0A0K8WJF4
Pubmed
19121390
11060215
26354079
26227816
22118469
12364791
+ More
25244985 17994087 18362917 19820115 25830018 26108605 25315136 17550304 18057021 20075255 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1334137 10876650 8917579 12537569 11517334 11861562 11880499 17893096 20798317 28004739 15632085 23185243 24508170 30249741 24438588 21347285 20920257 23761445 17510324 20966253 21719571 26483478 21282665 28648823
25244985 17994087 18362917 19820115 25830018 26108605 25315136 17550304 18057021 20075255 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1334137 10876650 8917579 12537569 11517334 11861562 11880499 17893096 20798317 28004739 15632085 23185243 24508170 30249741 24438588 21347285 20920257 23761445 17510324 20966253 21719571 26483478 21282665 28648823
EMBL
BABH01030871
BABH01030872
AF032873
AAF21642.1
ODYU01002761
SOQ40654.1
+ More
KQ460398 KPJ15092.1 KQ459606 KPI91475.1 JTDY01000103 KOB78852.1 NWSH01000473 PCG76156.1 AGBW02009960 OWR49499.1 GFDF01007312 JAV06772.1 AAAB01008984 GGFL01000921 MBW65099.1 GGFL01000922 MBW65100.1 GANO01003971 JAB55900.1 GFDL01003843 JAV31202.1 GFDL01005124 JAV29921.1 GFDL01005104 JAV29941.1 CH940649 EDW64292.2 KQ971372 EFA09465.2 GBXI01007616 JAD06676.1 CH963852 EDW75561.2 KA647302 AFP61931.1 JRES01000827 KNC28025.1 CM000157 EDW87651.1 CH902624 EDV33345.1 KPU74417.1 CM002910 KMY88097.1 CH480820 EDW54250.1 CH954177 EDV57745.2 CH933807 EDW12833.2 KRG03437.1 GDHF01022596 JAI29718.1 AE014134 ACZ94162.1 Y08362 U66460 AY128432 BT009955 AAC47292.1 AAF51001.1 AAM75025.1 AAQ22424.1 CAA69649.1 GL448255 EFN85026.1 GEZM01063070 JAV69333.1 OUUW01000006 SPP81406.1 CH479187 EDW39820.1 CH379060 EAL34145.2 KRT04538.1 CH916372 EDV99248.1 KK107144 QOIP01000003 EZA57571.1 RLU24820.1 APCN01003386 GL437616 EFN70243.1 KQ762842 OAD55412.1 ATLV01019462 KE525272 KFB44192.1 KZ288251 PBC31055.1 KQ976526 KYM81956.1 AXCM01004211 KQ976818 KYN08080.1 JXJN01024742 KC713793 AHH29251.1 KQ980155 KYN17462.1 AXCN02000741 ADTU01018181 ADTU01018182 ADMH02002027 ETN59874.1 AY314994 CH477389 AAP88701.1 EAT42015.1 GL888812 EGI57991.1 GFDL01005126 JAV29919.1 GFDL01005119 JAV29926.1 EAA14797.4 DS231876 EDS41983.1 KQ981424 KYN41855.1 JXUM01008641 JXUM01008642 JXUM01008643 KQ560265 KXJ83471.1 CVRI01000063 CRL04725.1 KRF81546.1 GL762903 EFZ20541.1 NNAY01000425 OXU28498.1 KQ434835 KZC07903.1 UFQT01001069 SSX28740.1 KRJ97284.1 CP012523 ALC38115.1 GEDC01021509 JAS15789.1 KPU74416.1 KQ982482 KYQ55914.1 KMY88095.1 KRG03439.1 KQS70099.1 GBXI01005671 JAD08621.1 GEBQ01015821 JAT24156.1 BT099793 ACV91630.1 SPP81407.1 GDHF01001038 JAI51276.1
KQ460398 KPJ15092.1 KQ459606 KPI91475.1 JTDY01000103 KOB78852.1 NWSH01000473 PCG76156.1 AGBW02009960 OWR49499.1 GFDF01007312 JAV06772.1 AAAB01008984 GGFL01000921 MBW65099.1 GGFL01000922 MBW65100.1 GANO01003971 JAB55900.1 GFDL01003843 JAV31202.1 GFDL01005124 JAV29921.1 GFDL01005104 JAV29941.1 CH940649 EDW64292.2 KQ971372 EFA09465.2 GBXI01007616 JAD06676.1 CH963852 EDW75561.2 KA647302 AFP61931.1 JRES01000827 KNC28025.1 CM000157 EDW87651.1 CH902624 EDV33345.1 KPU74417.1 CM002910 KMY88097.1 CH480820 EDW54250.1 CH954177 EDV57745.2 CH933807 EDW12833.2 KRG03437.1 GDHF01022596 JAI29718.1 AE014134 ACZ94162.1 Y08362 U66460 AY128432 BT009955 AAC47292.1 AAF51001.1 AAM75025.1 AAQ22424.1 CAA69649.1 GL448255 EFN85026.1 GEZM01063070 JAV69333.1 OUUW01000006 SPP81406.1 CH479187 EDW39820.1 CH379060 EAL34145.2 KRT04538.1 CH916372 EDV99248.1 KK107144 QOIP01000003 EZA57571.1 RLU24820.1 APCN01003386 GL437616 EFN70243.1 KQ762842 OAD55412.1 ATLV01019462 KE525272 KFB44192.1 KZ288251 PBC31055.1 KQ976526 KYM81956.1 AXCM01004211 KQ976818 KYN08080.1 JXJN01024742 KC713793 AHH29251.1 KQ980155 KYN17462.1 AXCN02000741 ADTU01018181 ADTU01018182 ADMH02002027 ETN59874.1 AY314994 CH477389 AAP88701.1 EAT42015.1 GL888812 EGI57991.1 GFDL01005126 JAV29919.1 GFDL01005119 JAV29926.1 EAA14797.4 DS231876 EDS41983.1 KQ981424 KYN41855.1 JXUM01008641 JXUM01008642 JXUM01008643 KQ560265 KXJ83471.1 CVRI01000063 CRL04725.1 KRF81546.1 GL762903 EFZ20541.1 NNAY01000425 OXU28498.1 KQ434835 KZC07903.1 UFQT01001069 SSX28740.1 KRJ97284.1 CP012523 ALC38115.1 GEDC01021509 JAS15789.1 KPU74416.1 KQ982482 KYQ55914.1 KMY88095.1 KRG03439.1 KQS70099.1 GBXI01005671 JAD08621.1 GEBQ01015821 JAT24156.1 BT099793 ACV91630.1 SPP81407.1 GDHF01001038 JAI51276.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000218220
UP000007151
+ More
UP000075920 UP000076408 UP000008792 UP000007266 UP000007798 UP000037069 UP000095300 UP000095301 UP000002282 UP000007801 UP000192221 UP000002358 UP000001292 UP000008711 UP000009192 UP000000803 UP000008237 UP000268350 UP000008744 UP000001819 UP000075885 UP000001070 UP000053097 UP000279307 UP000075840 UP000000311 UP000075884 UP000030765 UP000242457 UP000078200 UP000078540 UP000075883 UP000078542 UP000092443 UP000005203 UP000092460 UP000078492 UP000069272 UP000075886 UP000005205 UP000075880 UP000000673 UP000008820 UP000075882 UP000007755 UP000075903 UP000007062 UP000002320 UP000078541 UP000069940 UP000249989 UP000092445 UP000183832 UP000215335 UP000076502 UP000092553 UP000075809
UP000075920 UP000076408 UP000008792 UP000007266 UP000007798 UP000037069 UP000095300 UP000095301 UP000002282 UP000007801 UP000192221 UP000002358 UP000001292 UP000008711 UP000009192 UP000000803 UP000008237 UP000268350 UP000008744 UP000001819 UP000075885 UP000001070 UP000053097 UP000279307 UP000075840 UP000000311 UP000075884 UP000030765 UP000242457 UP000078200 UP000078540 UP000075883 UP000078542 UP000092443 UP000005203 UP000092460 UP000078492 UP000069272 UP000075886 UP000005205 UP000075880 UP000000673 UP000008820 UP000075882 UP000007755 UP000075903 UP000007062 UP000002320 UP000078541 UP000069940 UP000249989 UP000092445 UP000183832 UP000215335 UP000076502 UP000092553 UP000075809
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JBG2
Q9U5A5
A0A2H1VK69
A0A194RBW5
A0A194PK53
A0A0L7LTT3
+ More
A0A2A4JWJ5 A0A212F710 A0A1L8DJZ1 A0A182WG85 A0A1S4GZP7 A0A182Y0E7 A0A2M4CIM3 A0A2M4CIC0 U5EUA5 A0A1Q3FUS3 A0A1Q3FQM0 A0A1Q3FQP0 B4LUW5 D6X2M1 A0A0A1X708 B4MTX2 T1PET9 A0A0L0C6X9 A0A1I8NQS7 A0A1I8MTT9 B4NYY7 B3MU16 A0A1W4WA05 K7J4G0 A0A0J9QVL8 A0A1I8NQS8 B4I3A5 B3N3T6 B4KLF2 A0A0K8UTA4 E1JHT2 Q9VR07 E2BGZ3 A0A1Y1L6M7 A0A3B0K6Z8 B4GQE8 Q29LA0 A0A182P1M5 B4JQR8 A0A026WPJ4 A0A182I7E3 E2A8H3 A0A182NUS7 A0A310SJ50 A0A084W1U8 A0A2A3EH09 A0A1A9VW31 A0A195BBM1 A0A1S4GZP4 A0A182LUR7 A0A195D5A3 A0A1A9XJP4 A0A088A4T6 A0A1B0C304 X2D4U8 A0A195DXM1 A0A182F1I2 A0A182QL93 A0A158NJT5 A0A182J770 W5JAR1 Q7YZC7 A0A182LMQ1 F4X6F1 A0A1Q3FQS7 A0A182UTC3 A0A1Q3FR06 Q7PWJ5 B0WB25 A0A195FMG1 A0A182G8W1 A0A1A9ZCS2 A0A1J1IWZ2 A0A0Q9WA98 E9IFV1 A0A232FDE8 A0A154P7S6 A0A336MFN3 A0A0R1DJ70 A0A0M4E6D1 A0A1B6CQI5 A0A0P9A7T6 A0A1W4VY60 A0A1I8M2G9 A0A151X6F1 A0A0J9TFF7 A0A0Q9X5I5 A0A0Q5W9A0 A0A0A1XCF1 A0A1B6LKH7 Q9VR07-2 C8VV47 A0A3B0JGD1 A0A0K8WJF4
A0A2A4JWJ5 A0A212F710 A0A1L8DJZ1 A0A182WG85 A0A1S4GZP7 A0A182Y0E7 A0A2M4CIM3 A0A2M4CIC0 U5EUA5 A0A1Q3FUS3 A0A1Q3FQM0 A0A1Q3FQP0 B4LUW5 D6X2M1 A0A0A1X708 B4MTX2 T1PET9 A0A0L0C6X9 A0A1I8NQS7 A0A1I8MTT9 B4NYY7 B3MU16 A0A1W4WA05 K7J4G0 A0A0J9QVL8 A0A1I8NQS8 B4I3A5 B3N3T6 B4KLF2 A0A0K8UTA4 E1JHT2 Q9VR07 E2BGZ3 A0A1Y1L6M7 A0A3B0K6Z8 B4GQE8 Q29LA0 A0A182P1M5 B4JQR8 A0A026WPJ4 A0A182I7E3 E2A8H3 A0A182NUS7 A0A310SJ50 A0A084W1U8 A0A2A3EH09 A0A1A9VW31 A0A195BBM1 A0A1S4GZP4 A0A182LUR7 A0A195D5A3 A0A1A9XJP4 A0A088A4T6 A0A1B0C304 X2D4U8 A0A195DXM1 A0A182F1I2 A0A182QL93 A0A158NJT5 A0A182J770 W5JAR1 Q7YZC7 A0A182LMQ1 F4X6F1 A0A1Q3FQS7 A0A182UTC3 A0A1Q3FR06 Q7PWJ5 B0WB25 A0A195FMG1 A0A182G8W1 A0A1A9ZCS2 A0A1J1IWZ2 A0A0Q9WA98 E9IFV1 A0A232FDE8 A0A154P7S6 A0A336MFN3 A0A0R1DJ70 A0A0M4E6D1 A0A1B6CQI5 A0A0P9A7T6 A0A1W4VY60 A0A1I8M2G9 A0A151X6F1 A0A0J9TFF7 A0A0Q9X5I5 A0A0Q5W9A0 A0A0A1XCF1 A0A1B6LKH7 Q9VR07-2 C8VV47 A0A3B0JGD1 A0A0K8WJF4
PDB
4XPF
E-value=8.21111e-117,
Score=1078
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
Length:
727
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
267.8359
Exp number, first 60 AAs:
0
Total prob of N-in:
0.98126
inside
1 - 171
TMhelix
172 - 194
outside
195 - 203
TMhelix
204 - 226
inside
227 - 246
TMhelix
247 - 269
outside
270 - 330
TMhelix
331 - 353
inside
354 - 361
TMhelix
362 - 384
outside
385 - 410
TMhelix
411 - 433
inside
434 - 445
TMhelix
446 - 468
outside
469 - 505
TMhelix
506 - 528
inside
529 - 547
TMhelix
548 - 570
outside
571 - 579
TMhelix
580 - 602
inside
603 - 622
TMhelix
623 - 645
outside
646 - 659
TMhelix
660 - 682
inside
683 - 727
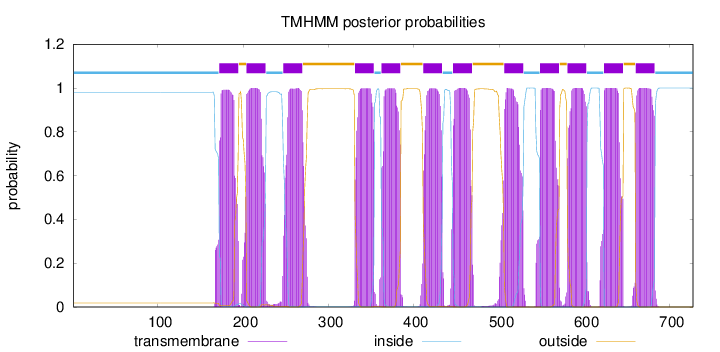
Population Genetic Test Statistics
Pi
240.272545
Theta
172.800898
Tajima's D
1.115228
CLR
0.01149
CSRT
0.686865656717164
Interpretation
Uncertain
