Pre Gene Modal
BGIBMGA006855
Annotation
PREDICTED:_dehydrogenase/reductase_SDR_family_protein_7-like_[Amyelois_transitella]
Full name
Dehydrogenase/reductase SDR family protein 7-like
Location in the cell
Mitochondrial Reliability : 2.412
Sequence
CDS
ATGTACGGGGTAATTAATCATCTTCTTCAAAAGAAAAGGAGATCAACGTTACGAGGAAAGGTGGTCTTAATTACTGGTGCAAGCTCTGGTATTGGGGAGGCACTGGCTCATGTATTTTATGGACAGGGCTGTAAAATTATCCTTGCTTCTCGGCGGAAAAGAGAACTCGAAAGAGTTAAACAGGATCTGATATCCAGGAAAGTTGCCATCCAAACTCTTGAACCAGTTGTTTTGGAGTTAAATTTAGCGGACATAGATAATATGTATTTGTTTGTGGAGAAAGTGTACGGGATATGTGGTCACGTTGACATTTTAGTCAATAATGGCGGTGTCACTCATAGAGGATCTATTTTAAACACTAAAACTGATGTTGATCAGAAAATTATGTTTATCAATTACTTCGGGTCAGTAGCATTGACAAAGGCTGTACTACCGAAAATGATAGAAAGAAAATCTGGGCATGTCGTATTTAATAGTTCTGTGCAAGGACTAATTTCCATACCTAATCGTTCAGCTTATGCTGCATCCAAGCATGCTCTACAAGCTTTTGGTGACTGTCTTCGTGCTGAGATGGATCAGTACGGTATTAATGTTACTGTTGTTAGCCCAGGGTATATAAAAACAGCTATATCAATGAACGCTTTCACCGGATCTGGTGACACTCATGGTGTTATGGATGCCACCACAGCGAAAGGTTTCACACCGGAATATACAGCGAACAAAATATTAGACGCGGTTGTTAATAAAAAAAATGAACTAATACTAAGCCAGTTTCTACCTAGATTTGCCATATTTTTGAGACACACATTTCCATCATTGTATTTTACGATTATGGCAAAAAGAGCGAACGAAACCTGA
Protein
MYGVINHLLQKKRRSTLRGKVVLITGASSGIGEALAHVFYGQGCKIILASRRKRELERVKQDLISRKVAIQTLEPVVLELNLADIDNMYLFVEKVYGICGHVDILVNNGGVTHRGSILNTKTDVDQKIMFINYFGSVALTKAVLPKMIERKSGHVVFNSSVQGLISIPNRSAYAASKHALQAFGDCLRAEMDQYGINVTVVSPGYIKTAISMNAFTGSGDTHGVMDATTAKGFTPEYTANKILDAVVNKKNELILSQFLPRFAIFLRHTFPSLYFTIMAKRANET
Summary
Description
Putative oxidoreductase.
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Keywords
Complete proteome
Membrane
NAD
NADP
Oxidoreductase
Peroxisome
Reference proteome
Signal-anchor
Transmembrane
Transmembrane helix
Feature
chain Dehydrogenase/reductase SDR family protein 7-like
Uniprot
H9JBG0
A0A2A4JUH6
S4P9S5
A0A2H1X2R5
A0A212EU93
A0A194RBX0
+ More
A0A194PK57 A0A0L7LK88 A0A1B6CN93 D6WJU2 A0A1W4WJT9 A0A088AVJ1 A0A0L7RIW1 B3P5E2 A0A1Y1LQ13 N6T7U5 U4UAG6 A0A0T6B6M8 A0A067QPU3 A0A158NCI8 A0A2A3ERB5 V9IC57 E2A2P1 B3MTH9 A0A1B0EZA9 A0A1W4UD14 B4PPL2 A0A182PP56 B4R079 E9IM84 A0A151X5T4 A0A3B0KGV6 B4K608 A0A1B0BNY4 A0A1A9XMV5 A0A182Y100 A0A182WKC7 Q9Y140 A0A182RN17 A0A195BYF9 A0A182LU11 A0A1S3HFH7 A0A182G9X2 A0A182QAV2 F4WX49 A0A023EPZ5 A0A1A9UQ71 B4HZD7 A0A0A1WFF3 Q29BT9 B4GPN4 A0A195CR11 A0A0M5JD48 A0A182NM29 A0A084VZ77 A0A182SM19 A0A1B0A8W7 W8BL68 A0A0K8U0G4 B4M5S7 A0A182UNJ8 A0A182JWG1 A0A1J1IPE9 A0A2M4AY01 A0A182X550 B0WPM0 Q7Q732 A0A195EMR5 A0A034WH73 A0A1B6EUP1 B4JYM2 A0A182J910 A0A2M4BWN2 A0A182FHF8 W5JU45 A0A182HLZ9 A0A2R2MQJ0 A0A0K8TSL6 A0A1S3HIF6 K7JAN2 J9JV73 A0A232EMU9 U5EYE3 A0A2S2NLR2 E2BU12 A0A1A9W6J2 A0A195F9D1 A0A2H8TUL4 A0A2R5L9W9 A0A1I8PCD4 B4NBK7 A0A026WVT9 A0A0J7KQT0 A0A0L0BQG0 A0A3L8DHM6 A0A1I8MFF2 T1ITV2 A0A0A9WVM2 A0A1Q3F839 V5HUV6
A0A194PK57 A0A0L7LK88 A0A1B6CN93 D6WJU2 A0A1W4WJT9 A0A088AVJ1 A0A0L7RIW1 B3P5E2 A0A1Y1LQ13 N6T7U5 U4UAG6 A0A0T6B6M8 A0A067QPU3 A0A158NCI8 A0A2A3ERB5 V9IC57 E2A2P1 B3MTH9 A0A1B0EZA9 A0A1W4UD14 B4PPL2 A0A182PP56 B4R079 E9IM84 A0A151X5T4 A0A3B0KGV6 B4K608 A0A1B0BNY4 A0A1A9XMV5 A0A182Y100 A0A182WKC7 Q9Y140 A0A182RN17 A0A195BYF9 A0A182LU11 A0A1S3HFH7 A0A182G9X2 A0A182QAV2 F4WX49 A0A023EPZ5 A0A1A9UQ71 B4HZD7 A0A0A1WFF3 Q29BT9 B4GPN4 A0A195CR11 A0A0M5JD48 A0A182NM29 A0A084VZ77 A0A182SM19 A0A1B0A8W7 W8BL68 A0A0K8U0G4 B4M5S7 A0A182UNJ8 A0A182JWG1 A0A1J1IPE9 A0A2M4AY01 A0A182X550 B0WPM0 Q7Q732 A0A195EMR5 A0A034WH73 A0A1B6EUP1 B4JYM2 A0A182J910 A0A2M4BWN2 A0A182FHF8 W5JU45 A0A182HLZ9 A0A2R2MQJ0 A0A0K8TSL6 A0A1S3HIF6 K7JAN2 J9JV73 A0A232EMU9 U5EYE3 A0A2S2NLR2 E2BU12 A0A1A9W6J2 A0A195F9D1 A0A2H8TUL4 A0A2R5L9W9 A0A1I8PCD4 B4NBK7 A0A026WVT9 A0A0J7KQT0 A0A0L0BQG0 A0A3L8DHM6 A0A1I8MFF2 T1ITV2 A0A0A9WVM2 A0A1Q3F839 V5HUV6
EC Number
1.1.-.-
Pubmed
19121390
23622113
22118469
26354079
26227816
18362917
+ More
19820115 17994087 28004739 23537049 24845553 21347285 20798317 17550304 21282665 25244985 10731132 12537572 10731138 26483478 21719571 24945155 25830018 15632085 24438588 24495485 12364791 25348373 20920257 23761445 26369729 20075255 28648823 24508170 26108605 30249741 25315136 25401762 26823975 25765539
19820115 17994087 28004739 23537049 24845553 21347285 20798317 17550304 21282665 25244985 10731132 12537572 10731138 26483478 21719571 24945155 25830018 15632085 24438588 24495485 12364791 25348373 20920257 23761445 26369729 20075255 28648823 24508170 26108605 30249741 25315136 25401762 26823975 25765539
EMBL
BABH01030861
NWSH01000646
PCG75052.1
GAIX01003609
JAA88951.1
ODYU01012985
+ More
SOQ59567.1 AGBW02012455 OWR45027.1 KQ460398 KPJ15097.1 KQ459606 KPI91480.1 JTDY01000870 KOB75606.1 GEDC01022351 JAS14947.1 KQ971343 EFA03099.1 KQ414582 KOC70882.1 CH954182 EDV53192.1 GEZM01049898 JAV75774.1 APGK01040850 KB740984 ENN76299.1 KB631899 ERL86945.1 LJIG01009469 KRT83018.1 KK853080 KDR11690.1 ADTU01011939 KZ288198 PBC33669.1 JR039437 AEY58703.1 GL436181 EFN72306.1 CH902623 EDV30569.1 AJVK01033643 CM000160 EDW98262.1 CM000364 EDX14885.1 GL764129 EFZ18513.1 KQ982490 KYQ55773.1 OUUW01000007 SPP82908.1 CH933806 EDW16245.1 JXJN01017678 AE014297 AF145631 KQ976396 KYM92973.1 AXCM01003247 JXUM01049886 KQ561625 KXJ77964.1 AXCN02002327 GL888417 EGI61292.1 GAPW01002470 JAC11128.1 CH480819 EDW53394.1 GBXI01016891 GBXI01016743 JAC97400.1 JAC97548.1 CM000070 EAL26908.2 CH479186 EDW39117.1 KQ977408 KYN02927.1 CP012526 ALC48056.1 ATLV01018617 KE525239 KFB43271.1 GAMC01012504 JAB94051.1 GDHF01032286 JAI20028.1 CH940652 EDW59003.2 CVRI01000054 CRL00369.1 GGFK01012358 MBW45679.1 DS232027 EDS32423.1 AAAB01008960 KQ978691 KYN29187.1 GAKP01005293 GAKP01005290 JAC53662.1 GECZ01028139 JAS41630.1 CH916377 EDV90784.1 GGFJ01008302 MBW57443.1 ADMH02000483 ETN66284.1 APCN01004290 GDAI01000234 JAI17369.1 AAZX01001115 AAZX01001121 AAZX01004158 AAZX01016586 ABLF02019394 NNAY01003277 OXU19694.1 GANO01002052 JAB57819.1 GGMR01005504 MBY18123.1 GL450552 EFN80815.1 KQ981727 KYN36986.1 GFXV01005696 MBW17501.1 GGLE01002152 MBY06278.1 CH964232 EDW81171.1 KK107088 EZA59786.1 LBMM01004296 KMQ92594.1 JRES01001623 KNC21444.1 QOIP01000008 RLU19419.1 JH431501 GBHO01031806 GBHO01031805 GBHO01031804 GBRD01014039 GBRD01014038 GBRD01014037 GBRD01014036 GDHC01010280 JAG11798.1 JAG11799.1 JAG11800.1 JAG51787.1 JAQ08349.1 GFDL01011310 JAV23735.1 GANP01006360 JAB78108.1
SOQ59567.1 AGBW02012455 OWR45027.1 KQ460398 KPJ15097.1 KQ459606 KPI91480.1 JTDY01000870 KOB75606.1 GEDC01022351 JAS14947.1 KQ971343 EFA03099.1 KQ414582 KOC70882.1 CH954182 EDV53192.1 GEZM01049898 JAV75774.1 APGK01040850 KB740984 ENN76299.1 KB631899 ERL86945.1 LJIG01009469 KRT83018.1 KK853080 KDR11690.1 ADTU01011939 KZ288198 PBC33669.1 JR039437 AEY58703.1 GL436181 EFN72306.1 CH902623 EDV30569.1 AJVK01033643 CM000160 EDW98262.1 CM000364 EDX14885.1 GL764129 EFZ18513.1 KQ982490 KYQ55773.1 OUUW01000007 SPP82908.1 CH933806 EDW16245.1 JXJN01017678 AE014297 AF145631 KQ976396 KYM92973.1 AXCM01003247 JXUM01049886 KQ561625 KXJ77964.1 AXCN02002327 GL888417 EGI61292.1 GAPW01002470 JAC11128.1 CH480819 EDW53394.1 GBXI01016891 GBXI01016743 JAC97400.1 JAC97548.1 CM000070 EAL26908.2 CH479186 EDW39117.1 KQ977408 KYN02927.1 CP012526 ALC48056.1 ATLV01018617 KE525239 KFB43271.1 GAMC01012504 JAB94051.1 GDHF01032286 JAI20028.1 CH940652 EDW59003.2 CVRI01000054 CRL00369.1 GGFK01012358 MBW45679.1 DS232027 EDS32423.1 AAAB01008960 KQ978691 KYN29187.1 GAKP01005293 GAKP01005290 JAC53662.1 GECZ01028139 JAS41630.1 CH916377 EDV90784.1 GGFJ01008302 MBW57443.1 ADMH02000483 ETN66284.1 APCN01004290 GDAI01000234 JAI17369.1 AAZX01001115 AAZX01001121 AAZX01004158 AAZX01016586 ABLF02019394 NNAY01003277 OXU19694.1 GANO01002052 JAB57819.1 GGMR01005504 MBY18123.1 GL450552 EFN80815.1 KQ981727 KYN36986.1 GFXV01005696 MBW17501.1 GGLE01002152 MBY06278.1 CH964232 EDW81171.1 KK107088 EZA59786.1 LBMM01004296 KMQ92594.1 JRES01001623 KNC21444.1 QOIP01000008 RLU19419.1 JH431501 GBHO01031806 GBHO01031805 GBHO01031804 GBRD01014039 GBRD01014038 GBRD01014037 GBRD01014036 GDHC01010280 JAG11798.1 JAG11799.1 JAG11800.1 JAG51787.1 JAQ08349.1 GFDL01011310 JAV23735.1 GANP01006360 JAB78108.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000007266 UP000192223 UP000005203 UP000053825 UP000008711 UP000019118 UP000030742 UP000027135 UP000005205 UP000242457 UP000000311 UP000007801 UP000092462 UP000192221 UP000002282 UP000075885 UP000000304 UP000075809 UP000268350 UP000009192 UP000092460 UP000092443 UP000076408 UP000075920 UP000000803 UP000075900 UP000078540 UP000075883 UP000085678 UP000069940 UP000249989 UP000075886 UP000007755 UP000078200 UP000001292 UP000001819 UP000008744 UP000078542 UP000092553 UP000075884 UP000030765 UP000075901 UP000092445 UP000008792 UP000075903 UP000075881 UP000183832 UP000076407 UP000002320 UP000007062 UP000078492 UP000001070 UP000075880 UP000069272 UP000000673 UP000075840 UP000002358 UP000007819 UP000215335 UP000008237 UP000091820 UP000078541 UP000095300 UP000007798 UP000053097 UP000036403 UP000037069 UP000279307 UP000095301
UP000007266 UP000192223 UP000005203 UP000053825 UP000008711 UP000019118 UP000030742 UP000027135 UP000005205 UP000242457 UP000000311 UP000007801 UP000092462 UP000192221 UP000002282 UP000075885 UP000000304 UP000075809 UP000268350 UP000009192 UP000092460 UP000092443 UP000076408 UP000075920 UP000000803 UP000075900 UP000078540 UP000075883 UP000085678 UP000069940 UP000249989 UP000075886 UP000007755 UP000078200 UP000001292 UP000001819 UP000008744 UP000078542 UP000092553 UP000075884 UP000030765 UP000075901 UP000092445 UP000008792 UP000075903 UP000075881 UP000183832 UP000076407 UP000002320 UP000007062 UP000078492 UP000001070 UP000075880 UP000069272 UP000000673 UP000075840 UP000002358 UP000007819 UP000215335 UP000008237 UP000091820 UP000078541 UP000095300 UP000007798 UP000053097 UP000036403 UP000037069 UP000279307 UP000095301
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JBG0
A0A2A4JUH6
S4P9S5
A0A2H1X2R5
A0A212EU93
A0A194RBX0
+ More
A0A194PK57 A0A0L7LK88 A0A1B6CN93 D6WJU2 A0A1W4WJT9 A0A088AVJ1 A0A0L7RIW1 B3P5E2 A0A1Y1LQ13 N6T7U5 U4UAG6 A0A0T6B6M8 A0A067QPU3 A0A158NCI8 A0A2A3ERB5 V9IC57 E2A2P1 B3MTH9 A0A1B0EZA9 A0A1W4UD14 B4PPL2 A0A182PP56 B4R079 E9IM84 A0A151X5T4 A0A3B0KGV6 B4K608 A0A1B0BNY4 A0A1A9XMV5 A0A182Y100 A0A182WKC7 Q9Y140 A0A182RN17 A0A195BYF9 A0A182LU11 A0A1S3HFH7 A0A182G9X2 A0A182QAV2 F4WX49 A0A023EPZ5 A0A1A9UQ71 B4HZD7 A0A0A1WFF3 Q29BT9 B4GPN4 A0A195CR11 A0A0M5JD48 A0A182NM29 A0A084VZ77 A0A182SM19 A0A1B0A8W7 W8BL68 A0A0K8U0G4 B4M5S7 A0A182UNJ8 A0A182JWG1 A0A1J1IPE9 A0A2M4AY01 A0A182X550 B0WPM0 Q7Q732 A0A195EMR5 A0A034WH73 A0A1B6EUP1 B4JYM2 A0A182J910 A0A2M4BWN2 A0A182FHF8 W5JU45 A0A182HLZ9 A0A2R2MQJ0 A0A0K8TSL6 A0A1S3HIF6 K7JAN2 J9JV73 A0A232EMU9 U5EYE3 A0A2S2NLR2 E2BU12 A0A1A9W6J2 A0A195F9D1 A0A2H8TUL4 A0A2R5L9W9 A0A1I8PCD4 B4NBK7 A0A026WVT9 A0A0J7KQT0 A0A0L0BQG0 A0A3L8DHM6 A0A1I8MFF2 T1ITV2 A0A0A9WVM2 A0A1Q3F839 V5HUV6
A0A194PK57 A0A0L7LK88 A0A1B6CN93 D6WJU2 A0A1W4WJT9 A0A088AVJ1 A0A0L7RIW1 B3P5E2 A0A1Y1LQ13 N6T7U5 U4UAG6 A0A0T6B6M8 A0A067QPU3 A0A158NCI8 A0A2A3ERB5 V9IC57 E2A2P1 B3MTH9 A0A1B0EZA9 A0A1W4UD14 B4PPL2 A0A182PP56 B4R079 E9IM84 A0A151X5T4 A0A3B0KGV6 B4K608 A0A1B0BNY4 A0A1A9XMV5 A0A182Y100 A0A182WKC7 Q9Y140 A0A182RN17 A0A195BYF9 A0A182LU11 A0A1S3HFH7 A0A182G9X2 A0A182QAV2 F4WX49 A0A023EPZ5 A0A1A9UQ71 B4HZD7 A0A0A1WFF3 Q29BT9 B4GPN4 A0A195CR11 A0A0M5JD48 A0A182NM29 A0A084VZ77 A0A182SM19 A0A1B0A8W7 W8BL68 A0A0K8U0G4 B4M5S7 A0A182UNJ8 A0A182JWG1 A0A1J1IPE9 A0A2M4AY01 A0A182X550 B0WPM0 Q7Q732 A0A195EMR5 A0A034WH73 A0A1B6EUP1 B4JYM2 A0A182J910 A0A2M4BWN2 A0A182FHF8 W5JU45 A0A182HLZ9 A0A2R2MQJ0 A0A0K8TSL6 A0A1S3HIF6 K7JAN2 J9JV73 A0A232EMU9 U5EYE3 A0A2S2NLR2 E2BU12 A0A1A9W6J2 A0A195F9D1 A0A2H8TUL4 A0A2R5L9W9 A0A1I8PCD4 B4NBK7 A0A026WVT9 A0A0J7KQT0 A0A0L0BQG0 A0A3L8DHM6 A0A1I8MFF2 T1ITV2 A0A0A9WVM2 A0A1Q3F839 V5HUV6
PDB
3OEC
E-value=2.7351e-21,
Score=250
Ontologies
GO
Topology
Subcellular location
Peroxisome membrane
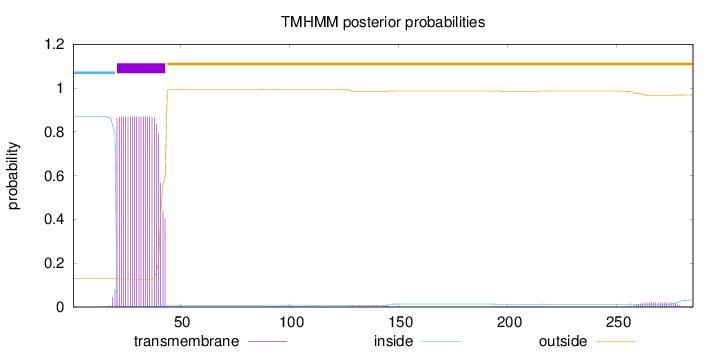
Length:
285
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.44175
Exp number, first 60 AAs:
18.81947
Total prob of N-in:
0.87058
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 285
Population Genetic Test Statistics
Pi
268.488145
Theta
196.127415
Tajima's D
0.133859
CLR
0.372229
CSRT
0.404079796010199
Interpretation
Uncertain
