Gene
KWMTBOMO05688 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006851
Annotation
PREDICTED:_splicing_factor_3B_subunit_1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.14 Mitochondrial Reliability : 1.314 Nuclear Reliability : 1.356
Sequence
CDS
ATGGATAAAATACCAAGAACTCATGAAGCCATTGAAGCACAAATAAAAGAAATACAATCAAAAAAAACTGAAGCACCAGAAAATGGTTCCAACAAAGGCGTGTCTCTCGGCGAGACTTACTATGACAGCGATATATATGATAGCACAGGCCAAGGAGGCAAGTCCAGATATGACGGATATGTGACATCAATTGCTGCTAATGACGAAGTAGAAGATGATGACCTCGAAAATGTTCCTATAACTCAGAAAAGGCCAGGATATACAGCACCTGCTTCACTGTTGAATGACATTGCACAAAGTGATAAAGAATTCGATCCTTTTGCTGACAAACGAAGACCAACTATCGCTGATAGAGAGGATGAATATCGACAAAAGAGGCGCAGGATGATTATATCACCTGAAAGAAATGATCCTTTTGCAGAAGGTGGGAAAACTCCTGATGTCGGATCGAGGACATACACTGATATAATGAAAGAGCAGTATCTTCTTGCTGAAGAAACAGAACTCAGAAAGAAACTGTTAGAGAAAGCTAGGGAAGGAACTCTCAAACCAGTACCACAATCTAATGGAGAATCCATAAAACCAGCAGCAAAACGCAAAGGTAGATGGGATCAAAGTTCTGAAGAAACACCAGTTGTTAAGAAAACAGTATCAACTACACCAAGCTCACAGACAACTCCGTCTTGGGATGCTGAGAGAGGCACATGGGAAGAAACTCCGGGAGCAGGTCGGGGCGGCGAGACGCCCGGTGCCACGCCATCGGCACGTGTTTGGGATGCCACACCAGGACATATCACTCCTGGACACGCCACTCCGGGGCGCGAAACTCCGGCTCATCACGCTTCTCGCAGAAATCGCTGGGATGAAACACCAAAAACAGATAGAGAAACACCAGGACACAACAGCGGTTGGGCTGAGACACCACGAACAGATCGCGGGTCAGGCGTGGATACAATACAGGAGACCCCCACCCCTGGCACAAAGCGGCGCTCGCGTTGGGACGAAACCCCCGGTGCCACGCCCGCACCCGCCACCCCCACGCCTTCTCACGCCACGCCTTCCCATGCCACACCCTCGCACGCCACGCCATCGCTCTCTACACCGACGCCGCACACGCCTGTCTTCACGCCAGGAGGGTCTACACCTGTGGGAGTGAAAGCCATGGCACTGGCTACACCAACACCTGGTCATGTTGCAGCCATGACTCCAGAACAGTTACAAGCTTATCGTTGGGAGAAAGAAATAGACGAGCGTAATAGACCTTACACTGATGAAGAACTGGATGCCATGTTCCCTCCTGGGTACAAGGTTTTACCACCGCCTGCCGGGTACGTCCCGATTCGGACGCCGGCCCGCAAGCTGACGGCGACGCCCACGCCGTTGGGTGGTACTCCAATCGGGTTTTTCATGCAGACGGAGGAGCCGGGCGGCTCGGCCGCGGCGGCCGCGCGTCTTCTCGACCCTCAGCCCAAGGGTAACCAGCAGCTGCCCTTTATGAAGCCCGAGGACGCCCAGTACTTCGACAAACTTCTCATCGACGTCGACGAGGATACACTCTCTCCCGACGAACTTAAGGAACGGAAAATAATGAAGCTTCTTTTGAAAATAAAGAACGGTACGCCACCCATGTGCAAAGCTGCCTTACGACAAATAACAGATAAAGCTCGCGAATTCGGTGCTGGACCACTTTTCAATCAAATTCTGCCATTACTGATGAGTCCTACGCTGGAAGATCAAGAGAGACATCTACTGGTCAAAGTCATAGACAGGATTTTGTATAAATTAGACGACTTGGTTCGACCCTACGTACATAAGATTCTTGTCGTCATTGAACCTCTGCTTATTGATGAAGATTATTATGCGCGTGTCGAAGGTCGCGAGATCATTTCAAATTTAGCTAAAGCAGCCGGCTTGGCTACTATGATTTCTACAATGAGGCCTGATATAGACAACATAGATGAGTATGTACGAAACACAACCGCTCGAGCGTTTGCGGTAGTGGCATCGGCCTTGGGTATTCCATCTTTATTGCCATTTTTAAAAGCTGTGTGCAGATCTAAGAAATCCTGGCAAGCACGTCACACAGGAATAAAGATTGTGCAACAAATTGCTATTCTGATGGGCTGTGCTATTCTGCCACATTTGAAATCTCTTGTTGAAATTATTGAACATGGTCTGGTGGACGAACAACAGAAGGTCCGAACGATCACGGCGCTGGCGAGCGCGGCTCTGGCCGAGGCCGCCACGCCCTACGGTATCGAGTCCTTCGATTCCGTGCTCAAACCTCTGTGGAAAGGTATTCGTACGCATCGTGGTAAAGGTCTGGCTGCATTCCTGAAAGCCATCGGATACCTCATCCCTCTCATGGATGCAGAATACGCGAATTACTACACACGAGAAGTGATGCTAATTCTTATTCGTGAGTTCCAGTCGCCCGATGAAGAAATGAAAAAAATCGTCTTAAAAGTAGTAAAACAATGTTGCGGTACCGACGGTGTTGAGCCTCAATACATAATGGATGAAATATTGCCTCATTTCTTTAAACACTTCTGGAACCATCGTATGGCCCTCGACAGACGAAACTATCGCCAGCTTGTCGACACCACAGTGGAAATTGCAAACAAGGTCGGTGCATCAGAAATTATCAACAGAATTGTTGATGATCTAAAAGACGACAATGAGCAGTATAGAAAAATGGTGATGGAGTCCATTGAAAAGATTCTCGCTAATTTAGGAGCAGCAGACATTGATTCTAAGCTCGAAGAAGCTCTTATCGATGGAATATTGTATGCATTCCAAGAACAGACAACTGAGGATGTTGTGATGCTGAACGGTTTTGGAACAATCGTGAATCAATTGGGAAAACGCGTAAAGCCCTATTTACCACAGATATGTGGTATTATTCTTTGGCGTATGAACAACAAGTCTGCTAAAGTTCGGCAACAAGCTGCCGATCTCATTTCTCGGATCGCTGTCGTGATGAAAACTTGTCAGGAGGAAAAGTTAATGGGGCATCTTGGTGTTGTACTGTATGAGTACTTGGGAGAGGAATATCCTGAAGTGCTAGGCTCTATACTTGGTGCATTAAAAGCAATTGTCAATGTTATTGGAATGACAAAGATGACCCCACCAATAAAGGATCTCCTGCCTAGGCTTACACCCATTTTGAAAAACAGGCATGAAAAAGTACAAGAGAATTGTATTGATTTAGTGGGTCGTATTGCAGACAGAGGACCGGAATTTGTGTCGGCCAGAGAATGGATGAGAATATGTTTTGAATTGCTTGAACTATTAAAGGCTCATAAGAAGGCAATTAGACGAGCGACTGTAAATACCTTTGGTTATATTGCTAAAGCCATCGGTCCTCATGACGTACTCGCAACCTTACTTAACAATCTGAAAGTGCAAGAGAGACAGAACAGAGTGTGCACAACGGTCGCTATTGCTATAGTTGCAGAGACCTGTTCACCTTTCACAGTGCTACCTGCTTTAATGAATGAATACAGAGTACCGGAATTAAATGTTCAAAACGGTGTGCTCAAATCACTATCTTTCCTTTTCGAGTACATTGGAGAAATGGGCAAGGATTACATTTATGCAGTTTGCCCATTATTGGAAGACGCGCTTATGGATCGAGATTTAGTACACAGACAAACGGCTTGCGCTGCCATAAAACATATGTCGCTCGGCGTGTATGGTTTCGGCTGTGAAGATGCCTTAGTACATCTATTAAATCATGTTTGGCCAAATATTTTTGAGACATCACCGCATCTGGTGCAAGCCTTCATGGACGCTGTTGAGGGCATGCGAGTTGCCCTAGGTCCCATTAAAATACTTCAATATGCATTACAGGGCCTGTTCCATCCAGCGAGGAAAGTACGCGACGTTTATTGGAAAATTTACAATACGCTTTACATCGGAGGACAGGACGCACTTGTTGCAGGATATCCCAGAATACAAAATGACCCAAATAATCACTTCATTAGATACGAATTAGATTATCTGCTTTAA
Protein
MDKIPRTHEAIEAQIKEIQSKKTEAPENGSNKGVSLGETYYDSDIYDSTGQGGKSRYDGYVTSIAANDEVEDDDLENVPITQKRPGYTAPASLLNDIAQSDKEFDPFADKRRPTIADREDEYRQKRRRMIISPERNDPFAEGGKTPDVGSRTYTDIMKEQYLLAEETELRKKLLEKAREGTLKPVPQSNGESIKPAAKRKGRWDQSSEETPVVKKTVSTTPSSQTTPSWDAERGTWEETPGAGRGGETPGATPSARVWDATPGHITPGHATPGRETPAHHASRRNRWDETPKTDRETPGHNSGWAETPRTDRGSGVDTIQETPTPGTKRRSRWDETPGATPAPATPTPSHATPSHATPSHATPSLSTPTPHTPVFTPGGSTPVGVKAMALATPTPGHVAAMTPEQLQAYRWEKEIDERNRPYTDEELDAMFPPGYKVLPPPAGYVPIRTPARKLTATPTPLGGTPIGFFMQTEEPGGSAAAAARLLDPQPKGNQQLPFMKPEDAQYFDKLLIDVDEDTLSPDELKERKIMKLLLKIKNGTPPMCKAALRQITDKAREFGAGPLFNQILPLLMSPTLEDQERHLLVKVIDRILYKLDDLVRPYVHKILVVIEPLLIDEDYYARVEGREIISNLAKAAGLATMISTMRPDIDNIDEYVRNTTARAFAVVASALGIPSLLPFLKAVCRSKKSWQARHTGIKIVQQIAILMGCAILPHLKSLVEIIEHGLVDEQQKVRTITALASAALAEAATPYGIESFDSVLKPLWKGIRTHRGKGLAAFLKAIGYLIPLMDAEYANYYTREVMLILIREFQSPDEEMKKIVLKVVKQCCGTDGVEPQYIMDEILPHFFKHFWNHRMALDRRNYRQLVDTTVEIANKVGASEIINRIVDDLKDDNEQYRKMVMESIEKILANLGAADIDSKLEEALIDGILYAFQEQTTEDVVMLNGFGTIVNQLGKRVKPYLPQICGIILWRMNNKSAKVRQQAADLISRIAVVMKTCQEEKLMGHLGVVLYEYLGEEYPEVLGSILGALKAIVNVIGMTKMTPPIKDLLPRLTPILKNRHEKVQENCIDLVGRIADRGPEFVSAREWMRICFELLELLKAHKKAIRRATVNTFGYIAKAIGPHDVLATLLNNLKVQERQNRVCTTVAIAIVAETCSPFTVLPALMNEYRVPELNVQNGVLKSLSFLFEYIGEMGKDYIYAVCPLLEDALMDRDLVHRQTACAAIKHMSLGVYGFGCEDALVHLLNHVWPNIFETSPHLVQAFMDAVEGMRVALGPIKILQYALQGLFHPARKVRDVYWKIYNTLYIGGQDALVAGYPRIQNDPNNHFIRYELDYLL
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. Trm1 family.
Uniprot
H9JBF6
A0A2A4JSP1
A0A194RFM2
A0A194PK67
A0A212EU62
A0A2H1VIF9
+ More
A0A1W4WWG8 A0A2J7R723 A0A154PJ75 A0A088AQ11 A0A146MA30 A0A0C9QR45 A0A2A3EIG8 A0A224X5B3 E0VK59 A0A1B6FM84 A0A232FLA7 K7IPV2 A0A0P4VU57 A0A139WC93 A0A0M8ZPW6 K7J1Y1 K7PQ80 A0A1Y1MRT7 B0WPC8 A0A1S4JNZ4 A0A2M4B9T2 W5JDY8 A0A182F0X8 A0A0L7QLT2 A0A2M4A9K8 T1I2N4 A0A182GWZ2 Q17F20 N6U455 E9IZA5 A0A1Q3FD79 A0A158P0S8 Q7QEW2 A0A195CXI4 A0A182P298 A0A3L8E186 U4UGU2 W4VRR9 A0A182YH49 A0A182MDD8 E2C8W2 A0A182N0F3 A0A182HIC7 F4WUU2 A0A151WWM8 A0A151JD02 A0A195FX49 U4U9S9 T1PIZ0 A0A182QVD5 A0A0K8VYL4 A0A1I8MGN7 A0A1B0GK95 A0A0L0BLC2 W8CEF0 A0A182Y565 A0A0A1X911 B4LTN6 B3MLP9 B4KFG6 B4JDR6 B4G880 B4P2G3 A0A0M5J8I6 A0A1L8DH50 B5DI06 A0A1J1HTB9 A0A1I8Q2K3 A0A195AUH1 B3N7S3 A0A3B0JBK8 Q9VPR5 B4N0T4 A0A1W4V410 A0A1B0DP60 A0A310SRE1 A0A0J9QTG0 B4ICZ1 E2AAN4 A0A1A9W934 A0A0J7KYV2 A0A1A9UHY6 A0A1A9Y8M1 A0A1B0C3V7 A0A1B0G5M0 A0A1A9ZBC0 A0A084WPU1 A0A182PUY4 A0A182IUQ0 T1IVP7 E9HF38 A0A182MHA2 L7M6K9 A0A224Z8I6
A0A1W4WWG8 A0A2J7R723 A0A154PJ75 A0A088AQ11 A0A146MA30 A0A0C9QR45 A0A2A3EIG8 A0A224X5B3 E0VK59 A0A1B6FM84 A0A232FLA7 K7IPV2 A0A0P4VU57 A0A139WC93 A0A0M8ZPW6 K7J1Y1 K7PQ80 A0A1Y1MRT7 B0WPC8 A0A1S4JNZ4 A0A2M4B9T2 W5JDY8 A0A182F0X8 A0A0L7QLT2 A0A2M4A9K8 T1I2N4 A0A182GWZ2 Q17F20 N6U455 E9IZA5 A0A1Q3FD79 A0A158P0S8 Q7QEW2 A0A195CXI4 A0A182P298 A0A3L8E186 U4UGU2 W4VRR9 A0A182YH49 A0A182MDD8 E2C8W2 A0A182N0F3 A0A182HIC7 F4WUU2 A0A151WWM8 A0A151JD02 A0A195FX49 U4U9S9 T1PIZ0 A0A182QVD5 A0A0K8VYL4 A0A1I8MGN7 A0A1B0GK95 A0A0L0BLC2 W8CEF0 A0A182Y565 A0A0A1X911 B4LTN6 B3MLP9 B4KFG6 B4JDR6 B4G880 B4P2G3 A0A0M5J8I6 A0A1L8DH50 B5DI06 A0A1J1HTB9 A0A1I8Q2K3 A0A195AUH1 B3N7S3 A0A3B0JBK8 Q9VPR5 B4N0T4 A0A1W4V410 A0A1B0DP60 A0A310SRE1 A0A0J9QTG0 B4ICZ1 E2AAN4 A0A1A9W934 A0A0J7KYV2 A0A1A9UHY6 A0A1A9Y8M1 A0A1B0C3V7 A0A1B0G5M0 A0A1A9ZBC0 A0A084WPU1 A0A182PUY4 A0A182IUQ0 T1IVP7 E9HF38 A0A182MHA2 L7M6K9 A0A224Z8I6
Pubmed
19121390
26354079
22118469
26823975
20566863
28648823
+ More
20075255 27129103 18362917 19820115 23139334 28004739 20920257 23761445 26483478 17510324 23537049 21282665 21347285 12364791 14747013 17210077 30249741 25244985 20798317 21719571 25315136 26108605 24495485 25830018 17994087 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24438588 21292972 25576852 28797301
20075255 27129103 18362917 19820115 23139334 28004739 20920257 23761445 26483478 17510324 23537049 21282665 21347285 12364791 14747013 17210077 30249741 25244985 20798317 21719571 25315136 26108605 24495485 25830018 17994087 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24438588 21292972 25576852 28797301
EMBL
BABH01030852
BABH01030853
BABH01030854
NWSH01000646
PCG75057.1
KQ460207
+ More
KPJ16623.1 KQ459606 KPI91490.1 AGBW02012455 OWR45033.1 ODYU01002544 SOQ40182.1 NEVH01006738 PNF36634.1 KQ434936 KZC11895.1 GDHC01003219 JAQ15410.1 GBYB01006194 JAG75961.1 KZ288232 PBC31498.1 GFTR01008741 JAW07685.1 DS235239 EEB13765.1 GECZ01018472 JAS51297.1 NNAY01000087 OXU31127.1 GDKW01001031 JAI55564.1 KQ971371 KYB25540.1 KQ435922 KOX68675.1 JX434608 AFP20535.1 GEZM01023516 JAV88341.1 DS232022 EDS32303.1 GGFJ01000652 MBW49793.1 ADMH02001427 ETN62577.1 KQ414905 KOC59608.1 GGFK01004162 MBW37483.1 ACPB03021045 JXUM01094257 JXUM01094258 KQ564116 KXJ72753.1 CH477277 EAT45112.1 APGK01050185 KB741167 ENN73372.1 GL767121 EFZ14106.1 GFDL01009576 JAV25469.1 ADTU01005778 AAAB01008846 EAA06480.5 KQ977141 KYN05373.1 QOIP01000001 RLU26386.1 KB632191 ERL89816.1 GANO01000389 JAB59482.1 AXCM01001335 GL453722 EFN75618.1 APCN01003927 GL888378 EGI62006.1 KQ982691 KYQ52065.1 KQ979038 KYN23473.1 KQ981200 KYN45001.1 ERL89817.1 KA648731 AFP63360.1 AXCN02001415 GDHF01008380 JAI43934.1 AJWK01028576 JRES01001701 KNC20842.1 GAMC01000311 JAC06245.1 GBXI01006675 GBXI01004113 JAD07617.1 JAD10179.1 CH940649 EDW65009.1 CH902620 EDV30770.1 CH933807 EDW13081.1 CH916368 EDW03436.1 CH479180 EDW28560.1 CM000157 EDW87159.1 CP012523 ALC37918.1 GFDF01008305 JAV05779.1 CH379060 EDY69943.1 CVRI01000020 CRK91312.1 KQ976738 KYM75670.1 CH954177 EDV57249.1 OUUW01000004 SPP79734.1 AE014134 AAF51478.2 AGB92374.1 CH963920 EDW77697.1 AJVK01007915 AJVK01007916 AJVK01007917 AJVK01007918 KQ761346 OAD57849.1 CM002910 KMY87341.1 CH480829 EDW45417.1 GL438137 EFN69484.1 LBMM01001954 KMQ95478.1 JXJN01017889 JXJN01025121 CCAG010013941 ATLV01025131 ATLV01025132 KE525369 KFB52235.1 AXCP01007967 JH431589 GL732633 EFX69656.1 AXCM01007375 GACK01005364 JAA59670.1 GFPF01012027 MAA23173.1
KPJ16623.1 KQ459606 KPI91490.1 AGBW02012455 OWR45033.1 ODYU01002544 SOQ40182.1 NEVH01006738 PNF36634.1 KQ434936 KZC11895.1 GDHC01003219 JAQ15410.1 GBYB01006194 JAG75961.1 KZ288232 PBC31498.1 GFTR01008741 JAW07685.1 DS235239 EEB13765.1 GECZ01018472 JAS51297.1 NNAY01000087 OXU31127.1 GDKW01001031 JAI55564.1 KQ971371 KYB25540.1 KQ435922 KOX68675.1 JX434608 AFP20535.1 GEZM01023516 JAV88341.1 DS232022 EDS32303.1 GGFJ01000652 MBW49793.1 ADMH02001427 ETN62577.1 KQ414905 KOC59608.1 GGFK01004162 MBW37483.1 ACPB03021045 JXUM01094257 JXUM01094258 KQ564116 KXJ72753.1 CH477277 EAT45112.1 APGK01050185 KB741167 ENN73372.1 GL767121 EFZ14106.1 GFDL01009576 JAV25469.1 ADTU01005778 AAAB01008846 EAA06480.5 KQ977141 KYN05373.1 QOIP01000001 RLU26386.1 KB632191 ERL89816.1 GANO01000389 JAB59482.1 AXCM01001335 GL453722 EFN75618.1 APCN01003927 GL888378 EGI62006.1 KQ982691 KYQ52065.1 KQ979038 KYN23473.1 KQ981200 KYN45001.1 ERL89817.1 KA648731 AFP63360.1 AXCN02001415 GDHF01008380 JAI43934.1 AJWK01028576 JRES01001701 KNC20842.1 GAMC01000311 JAC06245.1 GBXI01006675 GBXI01004113 JAD07617.1 JAD10179.1 CH940649 EDW65009.1 CH902620 EDV30770.1 CH933807 EDW13081.1 CH916368 EDW03436.1 CH479180 EDW28560.1 CM000157 EDW87159.1 CP012523 ALC37918.1 GFDF01008305 JAV05779.1 CH379060 EDY69943.1 CVRI01000020 CRK91312.1 KQ976738 KYM75670.1 CH954177 EDV57249.1 OUUW01000004 SPP79734.1 AE014134 AAF51478.2 AGB92374.1 CH963920 EDW77697.1 AJVK01007915 AJVK01007916 AJVK01007917 AJVK01007918 KQ761346 OAD57849.1 CM002910 KMY87341.1 CH480829 EDW45417.1 GL438137 EFN69484.1 LBMM01001954 KMQ95478.1 JXJN01017889 JXJN01025121 CCAG010013941 ATLV01025131 ATLV01025132 KE525369 KFB52235.1 AXCP01007967 JH431589 GL732633 EFX69656.1 AXCM01007375 GACK01005364 JAA59670.1 GFPF01012027 MAA23173.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000192223
+ More
UP000235965 UP000076502 UP000005203 UP000242457 UP000009046 UP000215335 UP000002358 UP000007266 UP000053105 UP000002320 UP000000673 UP000069272 UP000053825 UP000015103 UP000069940 UP000249989 UP000008820 UP000019118 UP000005205 UP000007062 UP000078542 UP000075885 UP000279307 UP000030742 UP000076408 UP000075883 UP000008237 UP000075884 UP000075840 UP000007755 UP000075809 UP000078492 UP000078541 UP000075886 UP000095301 UP000092461 UP000037069 UP000008792 UP000007801 UP000009192 UP000001070 UP000008744 UP000002282 UP000092553 UP000001819 UP000183832 UP000095300 UP000078540 UP000008711 UP000268350 UP000000803 UP000007798 UP000192221 UP000092462 UP000001292 UP000000311 UP000091820 UP000036403 UP000078200 UP000092443 UP000092460 UP000092444 UP000092445 UP000030765 UP000075880 UP000000305
UP000235965 UP000076502 UP000005203 UP000242457 UP000009046 UP000215335 UP000002358 UP000007266 UP000053105 UP000002320 UP000000673 UP000069272 UP000053825 UP000015103 UP000069940 UP000249989 UP000008820 UP000019118 UP000005205 UP000007062 UP000078542 UP000075885 UP000279307 UP000030742 UP000076408 UP000075883 UP000008237 UP000075884 UP000075840 UP000007755 UP000075809 UP000078492 UP000078541 UP000075886 UP000095301 UP000092461 UP000037069 UP000008792 UP000007801 UP000009192 UP000001070 UP000008744 UP000002282 UP000092553 UP000001819 UP000183832 UP000095300 UP000078540 UP000008711 UP000268350 UP000000803 UP000007798 UP000192221 UP000092462 UP000001292 UP000000311 UP000091820 UP000036403 UP000078200 UP000092443 UP000092460 UP000092444 UP000092445 UP000030765 UP000075880 UP000000305
Pfam
Interpro
IPR034085
TOG
+ More
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR038737 SF3b_su1-like
IPR015016 SF3b_su1
IPR021133 HEAT_type_2
IPR042296 tRNA_met_Trm1_C
IPR002905 Trm1
IPR022179 DUF3695
IPR029063 SAM-dependent_MTases
IPR017907 Znf_RING_CS
IPR032443 RAWUL
IPR005225 Small_GTP-bd_dom
IPR013083 Znf_RING/FYVE/PHD
IPR027417 P-loop_NTPase
IPR001841 Znf_RING
IPR001806 Small_GTPase
IPR022676 MyristoylCoA_TrFase_N
IPR016181 Acyl_CoA_acyltransferase
IPR022677 MyristoylCoA_TrFase_C
IPR022678 MyristoylCoA_TrFase_CS
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR038737 SF3b_su1-like
IPR015016 SF3b_su1
IPR021133 HEAT_type_2
IPR042296 tRNA_met_Trm1_C
IPR002905 Trm1
IPR022179 DUF3695
IPR029063 SAM-dependent_MTases
IPR017907 Znf_RING_CS
IPR032443 RAWUL
IPR005225 Small_GTP-bd_dom
IPR013083 Znf_RING/FYVE/PHD
IPR027417 P-loop_NTPase
IPR001841 Znf_RING
IPR001806 Small_GTPase
IPR022676 MyristoylCoA_TrFase_N
IPR016181 Acyl_CoA_acyltransferase
IPR022677 MyristoylCoA_TrFase_C
IPR022678 MyristoylCoA_TrFase_CS
Gene 3D
ProteinModelPortal
H9JBF6
A0A2A4JSP1
A0A194RFM2
A0A194PK67
A0A212EU62
A0A2H1VIF9
+ More
A0A1W4WWG8 A0A2J7R723 A0A154PJ75 A0A088AQ11 A0A146MA30 A0A0C9QR45 A0A2A3EIG8 A0A224X5B3 E0VK59 A0A1B6FM84 A0A232FLA7 K7IPV2 A0A0P4VU57 A0A139WC93 A0A0M8ZPW6 K7J1Y1 K7PQ80 A0A1Y1MRT7 B0WPC8 A0A1S4JNZ4 A0A2M4B9T2 W5JDY8 A0A182F0X8 A0A0L7QLT2 A0A2M4A9K8 T1I2N4 A0A182GWZ2 Q17F20 N6U455 E9IZA5 A0A1Q3FD79 A0A158P0S8 Q7QEW2 A0A195CXI4 A0A182P298 A0A3L8E186 U4UGU2 W4VRR9 A0A182YH49 A0A182MDD8 E2C8W2 A0A182N0F3 A0A182HIC7 F4WUU2 A0A151WWM8 A0A151JD02 A0A195FX49 U4U9S9 T1PIZ0 A0A182QVD5 A0A0K8VYL4 A0A1I8MGN7 A0A1B0GK95 A0A0L0BLC2 W8CEF0 A0A182Y565 A0A0A1X911 B4LTN6 B3MLP9 B4KFG6 B4JDR6 B4G880 B4P2G3 A0A0M5J8I6 A0A1L8DH50 B5DI06 A0A1J1HTB9 A0A1I8Q2K3 A0A195AUH1 B3N7S3 A0A3B0JBK8 Q9VPR5 B4N0T4 A0A1W4V410 A0A1B0DP60 A0A310SRE1 A0A0J9QTG0 B4ICZ1 E2AAN4 A0A1A9W934 A0A0J7KYV2 A0A1A9UHY6 A0A1A9Y8M1 A0A1B0C3V7 A0A1B0G5M0 A0A1A9ZBC0 A0A084WPU1 A0A182PUY4 A0A182IUQ0 T1IVP7 E9HF38 A0A182MHA2 L7M6K9 A0A224Z8I6
A0A1W4WWG8 A0A2J7R723 A0A154PJ75 A0A088AQ11 A0A146MA30 A0A0C9QR45 A0A2A3EIG8 A0A224X5B3 E0VK59 A0A1B6FM84 A0A232FLA7 K7IPV2 A0A0P4VU57 A0A139WC93 A0A0M8ZPW6 K7J1Y1 K7PQ80 A0A1Y1MRT7 B0WPC8 A0A1S4JNZ4 A0A2M4B9T2 W5JDY8 A0A182F0X8 A0A0L7QLT2 A0A2M4A9K8 T1I2N4 A0A182GWZ2 Q17F20 N6U455 E9IZA5 A0A1Q3FD79 A0A158P0S8 Q7QEW2 A0A195CXI4 A0A182P298 A0A3L8E186 U4UGU2 W4VRR9 A0A182YH49 A0A182MDD8 E2C8W2 A0A182N0F3 A0A182HIC7 F4WUU2 A0A151WWM8 A0A151JD02 A0A195FX49 U4U9S9 T1PIZ0 A0A182QVD5 A0A0K8VYL4 A0A1I8MGN7 A0A1B0GK95 A0A0L0BLC2 W8CEF0 A0A182Y565 A0A0A1X911 B4LTN6 B3MLP9 B4KFG6 B4JDR6 B4G880 B4P2G3 A0A0M5J8I6 A0A1L8DH50 B5DI06 A0A1J1HTB9 A0A1I8Q2K3 A0A195AUH1 B3N7S3 A0A3B0JBK8 Q9VPR5 B4N0T4 A0A1W4V410 A0A1B0DP60 A0A310SRE1 A0A0J9QTG0 B4ICZ1 E2AAN4 A0A1A9W934 A0A0J7KYV2 A0A1A9UHY6 A0A1A9Y8M1 A0A1B0C3V7 A0A1B0G5M0 A0A1A9ZBC0 A0A084WPU1 A0A182PUY4 A0A182IUQ0 T1IVP7 E9HF38 A0A182MHA2 L7M6K9 A0A224Z8I6
PDB
6QX9
E-value=0,
Score=5086
Ontologies
GO
PANTHER
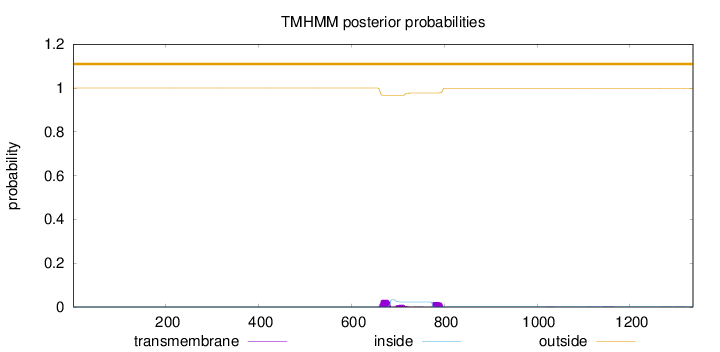
Topology
Length:
1336
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.50674000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00030
outside
1 - 1336
Population Genetic Test Statistics
Pi
280.906255
Theta
190.392468
Tajima's D
1.62928
CLR
0
CSRT
0.813209339533023
Interpretation
Uncertain
