Gene
KWMTBOMO05686
Annotation
PREDICTED:_EKC/KEOPS_complex_subunit_LAGE3_[Monomorium_pharaonis]
Location in the cell
Mitochondrial Reliability : 1.812
Sequence
CDS
ATGAACACGAAATCTACGGACAAAACAGCTATAACTCTAAAATTATCATTCACGTCCAATGATAATGCTTCTCTTGTCTACGATGTACTAAATGTTGACAAGGAATTAAAAGGAAGTGGTGTACATAGAGAATTTCAATTAAAGTCTAATGTTCTCTACATAGAATTTAAAAGCTTAGATTTAAAACGCCTAAGAGTAGCTGTGAATGCGATCCTAAAAAATATACTTCTTATCACAAAAACTGTGGAAAACTTTGCAACAAAATAA
Protein
MNTKSTDKTAITLKLSFTSNDNASLVYDVLNVDKELKGSGVHREFQLKSNVLYIEFKSLDLKRLRVAVNAILKNILLITKTVENFATK
Summary
Uniprot
A0A194QKK8
A0A0D2P650
A0A1S3KFB3
A0A165T744
A0A0P4WGF3
A0A0C3S8Q7
+ More
W8CCG3 A0A137QML4 A0A0C3C6V6 A0A0C2X5G3 A0A0C9MI60 E2LYG5 A0A1W4WZZ4 A0A158NK88 A0A2C9M747 A0A0W4ZWF5 V2XQB2 A0A0W0EXR4 K5WRW6 A0A1Y1Z8L1 A0A0B6ZCT3 A0A195BAY5 A0A0D7B9U0 A0A146IM92 A0A0D0CU53 A0A168L6I3 A0A1Q3G2M4 A0A1B0BF31 A0A397U743 A0A1Q3ESK4 V4A782 A0A1I8MCQ9 A0A0C3MKR9 A0A1I8QAD3 A0A1C7NET7 E2AVF3 A9P2A4 A0A139WHW3 A0A2P4QBL0 A0A2N1P2T6 A0A015K9E3 A0A168PCQ6 A0A367KT31 A0A1U8AG46 B0D9T1 F4W8I6 A0A151I7P0 A0A1R3JFI5 U9UE37 Q17FU1 A0A151X6X1 A0A372RW54 B7FFG5 A0A166G1G7 A0A164VLJ9 A0A2I1DT05 A0A1X0RQ38 A0A367KD54 A0A2H1VIV2 A0A0B7NXC0 A0A2N9IV80 A0A1U8A7D1 A0A0C9XCU4 B7FN15 A0A2H5MV06 A0A2I1GCT9 V7CVI7 A0A195EHR2 A0A3L5TRZ4 A0A177VE84 A0A1B0AA29 S8FJ38 K7J4Z2 C5XKY0 A0A2I4HMJ9 B3S784 A0A2Z6SM49 A0A1X2I7S9 A0A1L8EAH1 A0A1L8EAS0 A0A0B2RI12 I1LKK8 A0A1L8EAD7 A0A2H3EJV5 A0A1L8EAU1 B9S7G1 A0A2H5PXW6 A0A1L8EAP2 I1CTA7 A0A1X0QQN8 A0A058ZZR3 A0A232EYF9 A0A2P5RGT7 A0A1U8I3L3 A0A195FC84 S7PZ88 A0A371FIQ5 A0A2S3I468 A0A3L6RLY5 A0A2I4EYY5
W8CCG3 A0A137QML4 A0A0C3C6V6 A0A0C2X5G3 A0A0C9MI60 E2LYG5 A0A1W4WZZ4 A0A158NK88 A0A2C9M747 A0A0W4ZWF5 V2XQB2 A0A0W0EXR4 K5WRW6 A0A1Y1Z8L1 A0A0B6ZCT3 A0A195BAY5 A0A0D7B9U0 A0A146IM92 A0A0D0CU53 A0A168L6I3 A0A1Q3G2M4 A0A1B0BF31 A0A397U743 A0A1Q3ESK4 V4A782 A0A1I8MCQ9 A0A0C3MKR9 A0A1I8QAD3 A0A1C7NET7 E2AVF3 A9P2A4 A0A139WHW3 A0A2P4QBL0 A0A2N1P2T6 A0A015K9E3 A0A168PCQ6 A0A367KT31 A0A1U8AG46 B0D9T1 F4W8I6 A0A151I7P0 A0A1R3JFI5 U9UE37 Q17FU1 A0A151X6X1 A0A372RW54 B7FFG5 A0A166G1G7 A0A164VLJ9 A0A2I1DT05 A0A1X0RQ38 A0A367KD54 A0A2H1VIV2 A0A0B7NXC0 A0A2N9IV80 A0A1U8A7D1 A0A0C9XCU4 B7FN15 A0A2H5MV06 A0A2I1GCT9 V7CVI7 A0A195EHR2 A0A3L5TRZ4 A0A177VE84 A0A1B0AA29 S8FJ38 K7J4Z2 C5XKY0 A0A2I4HMJ9 B3S784 A0A2Z6SM49 A0A1X2I7S9 A0A1L8EAH1 A0A1L8EAS0 A0A0B2RI12 I1LKK8 A0A1L8EAD7 A0A2H3EJV5 A0A1L8EAU1 B9S7G1 A0A2H5PXW6 A0A1L8EAP2 I1CTA7 A0A1X0QQN8 A0A058ZZR3 A0A232EYF9 A0A2P5RGT7 A0A1U8I3L3 A0A195FC84 S7PZ88 A0A371FIQ5 A0A2S3I468 A0A3L6RLY5 A0A2I4EYY5
Pubmed
26354079
26659563
25474575
24495485
19019209
21347285
+ More
15562597 26899007 24571091 23045686 25683379 23254933 25315136 20798317 18854048 18362917 19820115 24277808 29355972 29674435 23663246 18322534 21719571 17510324 27956601 22089132 24767513 30397259 29259619 26977809 22745431 20075255 19189423 27145194 18719581 20075913 20729833 19578406 24919147 28648823 25893780 30683860
15562597 26899007 24571091 23045686 25683379 23254933 25315136 20798317 18854048 18362917 19820115 24277808 29355972 29674435 23663246 18322534 21719571 17510324 27956601 22089132 24767513 30397259 29259619 26977809 22745431 20075255 19189423 27145194 18719581 20075913 20729833 19578406 24919147 28648823 25893780 30683860
EMBL
KQ461198
KPJ06093.1
KN817528
KJA26399.1
KV429038
KZT73021.1
+ More
GDRN01011445 JAI67884.1 KN840493 KIP07617.1 GAMC01003746 GAMC01003744 JAC02810.1 KQ962155 KXN88502.1 KN831784 KIM39954.1 KN824278 KIM33378.1 DF836435 GAN07044.1 ABRE01017344 EEB89535.1 ADTU01018740 LFWA01000001 KTW32713.1 AWSO01000123 ESK94715.1 LATX01002462 KTB28836.1 JH971393 EKM78111.1 MCFE01000015 ORY06610.1 HACG01019468 CEK66333.1 KQ976537 KYM81354.1 KN880530 KIY67277.1 DF849967 GAT60689.1 KN834765 KIK63002.1 AMYB01000004 OAD03164.1 GFDL01000970 JAV34075.1 JXJN01013275 QKWP01001870 RIB06092.1 BDGU01001594 GAW10211.1 KB200907 ESO99803.1 KN822943 KIO34307.1 LUGH01000352 OBZ85844.1 GL443090 EFN62587.1 EF087782 ABK27015.1 KQ971343 KYB27484.1 AUPC02000065 POG75021.1 LLXL01000014 PKK80441.1 JEMT01012312 EXX76155.1 LT553674 SAM02156.1 PJQM01000410 RCI05355.1 DS547101 EDR08637.1 GL887908 EGI69477.1 KQ978406 KYM94166.1 AWUE01016254 OMO93589.1 KI279018 ESA18640.1 CH477268 EAT45410.1 KQ982472 KYQ56112.1 QKKE01000016 RGB42859.1 BT050686 BT135682 ACJ83355.1 AFK35477.1 KV428023 KZT41213.1 KV419405 KZS94263.1 LLXH01000020 LLXK01000016 PKC75569.1 PKY13020.1 KV921494 ORE14041.1 PJQL01000079 RCI00118.1 ODYU01002544 SOQ40184.1 LN734204 CEP20013.1 OIVN01006226 SPD28265.1 KN838536 KIK10055.1 BT053489 CM001217 BT147413 PSQE01000001 ACJ86148.1 AES58750.1 AFK47207.1 RHN76713.1 BDQV01001277 GAY31863.1 LLXJ01000098 LLXI01000321 PKC15198.1 PKY44421.1 CM002288 ESW33280.1 KQ978881 KYN27793.1 KV587457 OPL32703.1 LWDE01000704 OAJ26153.1 KE504142 EPT01441.1 CM000762 EES03676.1 DS985253 EDV21527.1 BEXD01004192 GBC08097.1 MCGE01000022 ORZ11200.1 GFDG01003071 JAV15728.1 GFDG01003070 JAV15729.1 KN650942 KHN31603.1 CM000850 KRH04984.1 GFDG01003069 JAV15730.1 KZ293644 PBL03573.1 GFDG01003074 JAV15725.1 EQ973885 EEF40454.1 BDQV01000156 GAY57217.1 GFDG01003075 JAV15724.1 CH476751 EIE91687.1 KV922076 ORE02059.1 KK198763 KCW46575.1 NNAY01001584 OXU23526.1 KZ647855 PPD86010.1 KQ981693 KYN37659.1 KB469307 EPQ52798.1 QJKJ01008936 RDX78197.1 CM008051 PAN36401.1 PQIB02000008 RLN05062.1
GDRN01011445 JAI67884.1 KN840493 KIP07617.1 GAMC01003746 GAMC01003744 JAC02810.1 KQ962155 KXN88502.1 KN831784 KIM39954.1 KN824278 KIM33378.1 DF836435 GAN07044.1 ABRE01017344 EEB89535.1 ADTU01018740 LFWA01000001 KTW32713.1 AWSO01000123 ESK94715.1 LATX01002462 KTB28836.1 JH971393 EKM78111.1 MCFE01000015 ORY06610.1 HACG01019468 CEK66333.1 KQ976537 KYM81354.1 KN880530 KIY67277.1 DF849967 GAT60689.1 KN834765 KIK63002.1 AMYB01000004 OAD03164.1 GFDL01000970 JAV34075.1 JXJN01013275 QKWP01001870 RIB06092.1 BDGU01001594 GAW10211.1 KB200907 ESO99803.1 KN822943 KIO34307.1 LUGH01000352 OBZ85844.1 GL443090 EFN62587.1 EF087782 ABK27015.1 KQ971343 KYB27484.1 AUPC02000065 POG75021.1 LLXL01000014 PKK80441.1 JEMT01012312 EXX76155.1 LT553674 SAM02156.1 PJQM01000410 RCI05355.1 DS547101 EDR08637.1 GL887908 EGI69477.1 KQ978406 KYM94166.1 AWUE01016254 OMO93589.1 KI279018 ESA18640.1 CH477268 EAT45410.1 KQ982472 KYQ56112.1 QKKE01000016 RGB42859.1 BT050686 BT135682 ACJ83355.1 AFK35477.1 KV428023 KZT41213.1 KV419405 KZS94263.1 LLXH01000020 LLXK01000016 PKC75569.1 PKY13020.1 KV921494 ORE14041.1 PJQL01000079 RCI00118.1 ODYU01002544 SOQ40184.1 LN734204 CEP20013.1 OIVN01006226 SPD28265.1 KN838536 KIK10055.1 BT053489 CM001217 BT147413 PSQE01000001 ACJ86148.1 AES58750.1 AFK47207.1 RHN76713.1 BDQV01001277 GAY31863.1 LLXJ01000098 LLXI01000321 PKC15198.1 PKY44421.1 CM002288 ESW33280.1 KQ978881 KYN27793.1 KV587457 OPL32703.1 LWDE01000704 OAJ26153.1 KE504142 EPT01441.1 CM000762 EES03676.1 DS985253 EDV21527.1 BEXD01004192 GBC08097.1 MCGE01000022 ORZ11200.1 GFDG01003071 JAV15728.1 GFDG01003070 JAV15729.1 KN650942 KHN31603.1 CM000850 KRH04984.1 GFDG01003069 JAV15730.1 KZ293644 PBL03573.1 GFDG01003074 JAV15725.1 EQ973885 EEF40454.1 BDQV01000156 GAY57217.1 GFDG01003075 JAV15724.1 CH476751 EIE91687.1 KV922076 ORE02059.1 KK198763 KCW46575.1 NNAY01001584 OXU23526.1 KZ647855 PPD86010.1 KQ981693 KYN37659.1 KB469307 EPQ52798.1 QJKJ01008936 RDX78197.1 CM008051 PAN36401.1 PQIB02000008 RLN05062.1
Proteomes
UP000053240
UP000054270
UP000085678
UP000076727
UP000053257
UP000070249
+ More
UP000053424 UP000054097 UP000053815 UP000000741 UP000192223 UP000005205 UP000076420 UP000053447 UP000017559 UP000054988 UP000008493 UP000193498 UP000078540 UP000054007 UP000077051 UP000092460 UP000266673 UP000188533 UP000030746 UP000095301 UP000054248 UP000095300 UP000093000 UP000000311 UP000007266 UP000018888 UP000233469 UP000022910 UP000078561 UP000253551 UP000189703 UP000001194 UP000007755 UP000078542 UP000187203 UP000008820 UP000075809 UP000263633 UP000076798 UP000076722 UP000232688 UP000242381 UP000252139 UP000054107 UP000054477 UP000002051 UP000265566 UP000236630 UP000232722 UP000234323 UP000000226 UP000078492 UP000077684 UP000092445 UP000015241 UP000002358 UP000000768 UP000235220 UP000009022 UP000247702 UP000193560 UP000008827 UP000217790 UP000008311 UP000009138 UP000242414 UP000030711 UP000215335 UP000239050 UP000189702 UP000078541 UP000030669 UP000257109 UP000243499 UP000275267
UP000053424 UP000054097 UP000053815 UP000000741 UP000192223 UP000005205 UP000076420 UP000053447 UP000017559 UP000054988 UP000008493 UP000193498 UP000078540 UP000054007 UP000077051 UP000092460 UP000266673 UP000188533 UP000030746 UP000095301 UP000054248 UP000095300 UP000093000 UP000000311 UP000007266 UP000018888 UP000233469 UP000022910 UP000078561 UP000253551 UP000189703 UP000001194 UP000007755 UP000078542 UP000187203 UP000008820 UP000075809 UP000263633 UP000076798 UP000076722 UP000232688 UP000242381 UP000252139 UP000054107 UP000054477 UP000002051 UP000265566 UP000236630 UP000232722 UP000234323 UP000000226 UP000078492 UP000077684 UP000092445 UP000015241 UP000002358 UP000000768 UP000235220 UP000009022 UP000247702 UP000193560 UP000008827 UP000217790 UP000008311 UP000009138 UP000242414 UP000030711 UP000215335 UP000239050 UP000189702 UP000078541 UP000030669 UP000257109 UP000243499 UP000275267
Interpro
ProteinModelPortal
A0A194QKK8
A0A0D2P650
A0A1S3KFB3
A0A165T744
A0A0P4WGF3
A0A0C3S8Q7
+ More
W8CCG3 A0A137QML4 A0A0C3C6V6 A0A0C2X5G3 A0A0C9MI60 E2LYG5 A0A1W4WZZ4 A0A158NK88 A0A2C9M747 A0A0W4ZWF5 V2XQB2 A0A0W0EXR4 K5WRW6 A0A1Y1Z8L1 A0A0B6ZCT3 A0A195BAY5 A0A0D7B9U0 A0A146IM92 A0A0D0CU53 A0A168L6I3 A0A1Q3G2M4 A0A1B0BF31 A0A397U743 A0A1Q3ESK4 V4A782 A0A1I8MCQ9 A0A0C3MKR9 A0A1I8QAD3 A0A1C7NET7 E2AVF3 A9P2A4 A0A139WHW3 A0A2P4QBL0 A0A2N1P2T6 A0A015K9E3 A0A168PCQ6 A0A367KT31 A0A1U8AG46 B0D9T1 F4W8I6 A0A151I7P0 A0A1R3JFI5 U9UE37 Q17FU1 A0A151X6X1 A0A372RW54 B7FFG5 A0A166G1G7 A0A164VLJ9 A0A2I1DT05 A0A1X0RQ38 A0A367KD54 A0A2H1VIV2 A0A0B7NXC0 A0A2N9IV80 A0A1U8A7D1 A0A0C9XCU4 B7FN15 A0A2H5MV06 A0A2I1GCT9 V7CVI7 A0A195EHR2 A0A3L5TRZ4 A0A177VE84 A0A1B0AA29 S8FJ38 K7J4Z2 C5XKY0 A0A2I4HMJ9 B3S784 A0A2Z6SM49 A0A1X2I7S9 A0A1L8EAH1 A0A1L8EAS0 A0A0B2RI12 I1LKK8 A0A1L8EAD7 A0A2H3EJV5 A0A1L8EAU1 B9S7G1 A0A2H5PXW6 A0A1L8EAP2 I1CTA7 A0A1X0QQN8 A0A058ZZR3 A0A232EYF9 A0A2P5RGT7 A0A1U8I3L3 A0A195FC84 S7PZ88 A0A371FIQ5 A0A2S3I468 A0A3L6RLY5 A0A2I4EYY5
W8CCG3 A0A137QML4 A0A0C3C6V6 A0A0C2X5G3 A0A0C9MI60 E2LYG5 A0A1W4WZZ4 A0A158NK88 A0A2C9M747 A0A0W4ZWF5 V2XQB2 A0A0W0EXR4 K5WRW6 A0A1Y1Z8L1 A0A0B6ZCT3 A0A195BAY5 A0A0D7B9U0 A0A146IM92 A0A0D0CU53 A0A168L6I3 A0A1Q3G2M4 A0A1B0BF31 A0A397U743 A0A1Q3ESK4 V4A782 A0A1I8MCQ9 A0A0C3MKR9 A0A1I8QAD3 A0A1C7NET7 E2AVF3 A9P2A4 A0A139WHW3 A0A2P4QBL0 A0A2N1P2T6 A0A015K9E3 A0A168PCQ6 A0A367KT31 A0A1U8AG46 B0D9T1 F4W8I6 A0A151I7P0 A0A1R3JFI5 U9UE37 Q17FU1 A0A151X6X1 A0A372RW54 B7FFG5 A0A166G1G7 A0A164VLJ9 A0A2I1DT05 A0A1X0RQ38 A0A367KD54 A0A2H1VIV2 A0A0B7NXC0 A0A2N9IV80 A0A1U8A7D1 A0A0C9XCU4 B7FN15 A0A2H5MV06 A0A2I1GCT9 V7CVI7 A0A195EHR2 A0A3L5TRZ4 A0A177VE84 A0A1B0AA29 S8FJ38 K7J4Z2 C5XKY0 A0A2I4HMJ9 B3S784 A0A2Z6SM49 A0A1X2I7S9 A0A1L8EAH1 A0A1L8EAS0 A0A0B2RI12 I1LKK8 A0A1L8EAD7 A0A2H3EJV5 A0A1L8EAU1 B9S7G1 A0A2H5PXW6 A0A1L8EAP2 I1CTA7 A0A1X0QQN8 A0A058ZZR3 A0A232EYF9 A0A2P5RGT7 A0A1U8I3L3 A0A195FC84 S7PZ88 A0A371FIQ5 A0A2S3I468 A0A3L6RLY5 A0A2I4EYY5
PDB
4WXA
E-value=0.00167525,
Score=91
Ontologies
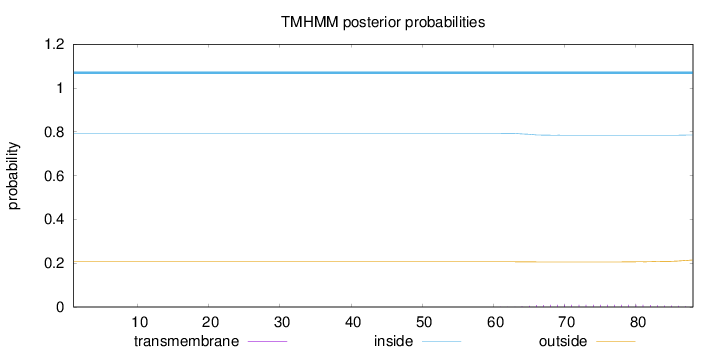
Topology
Length:
88
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2051
Exp number, first 60 AAs:
0.00101
Total prob of N-in:
0.79347
inside
1 - 88
Population Genetic Test Statistics
Pi
15.147527
Theta
22.640069
Tajima's D
-1.286048
CLR
0.577651
CSRT
0.0867456627168642
Interpretation
Uncertain
