Pre Gene Modal
BGIBMGA006849
Annotation
PREDICTED:_protein_RER1_[Amyelois_transitella]
Full name
Protein RER1
Location in the cell
PlasmaMembrane Reliability : 4.284
Sequence
CDS
ATGATGGACGGAAATGATTTATCTTCTGAAATTACAAGAAAAGGAATATTGTCTCAAGGTTGGACACGAATATCGCAGGTTTATCAGAGTACATTAGACAGATGGACCCCTCACGCAAAGACACGGTGGGTTGGAAGTGCCTTGGTCATGGCTGCATTTATTATCCGCATAATTACTAAACAAGGATGGTATATTGTCACATATGCGTTGGGTATTTACCATTTAAACTTGTTCATTGCATTCTTAACGCCGAAAATTGATCCTGCTATGGATTTAGATGATGATGAAAACGGACCTGCATTACCTACCAGAGCTAGTGAAGAATTTAGACCATTCATTAGACGGTTACCAGAATTCAAGTTTTGGCTGTCCGTTACCAAAAGCACTTTAATAGCTTTCTGCTGTACTTTTGTGGATGCTTTCAATATTCCAGTATTTTGGCCTATTCTAGTTATGTATTTCATAACATTATTTTGTATTACTATGAAGAGACAAATCAAGCATATGATCAAATACAGATATCTGCCATTTACACACAACAAACCAAAGTATAGAAATGCAGATTCTACAGTAACAGTGAACTGA
Protein
MMDGNDLSSEITRKGILSQGWTRISQVYQSTLDRWTPHAKTRWVGSALVMAAFIIRIITKQGWYIVTYALGIYHLNLFIAFLTPKIDPAMDLDDDENGPALPTRASEEFRPFIRRLPEFKFWLSVTKSTLIAFCCTFVDAFNIPVFWPILVMYFITLFCITMKRQIKHMIKYRYLPFTHNKPKYRNADSTVTVN
Summary
Description
Involved in the retrieval of endoplasmic reticulum membrane proteins from the early Golgi compartment.
Similarity
Belongs to the RER1 family.
Uniprot
A0A1E1WF01
A0A2A4K0S8
A0A194QMB3
I4DJJ4
A0A2H1VH83
S4PWX4
+ More
A0A3S2M9B6 A0A1W4X9J9 D6X1A4 A0A212EJL2 H9JBF4 A0A158NUW8 A0A195FPQ6 A0A3L8DFI4 A0A151WSV8 A0A336KQT1 A0A1W4X9B4 T1DF64 A0A151IRM5 A0A1Q3G3N1 A0A1Y1LLW8 A0A0K8TPZ6 Q16T78 A0A023EIL2 A0A182HD55 A0A1L8DUM1 A0A182RMQ7 A0A2M4AHY7 W5JML6 A0A182FVT0 A0A1J1INH3 T1E7W4 A0A2M3Z443 A0A182K8S4 A0A182QPE7 A0A154PEL8 A0A2M4AI32 A0A182N9Q2 A0A182YBQ4 F4X5Q3 A0A0L7QZ26 A0A2A3EN73 A0A0L0CIW1 A0A182VDM2 A0A182THS1 A0A182XFQ5 Q7PV51 A0A088ACJ9 A0A0M8ZX57 A0A195BVP6 V9II70 A0A067RF06 A0A0T6AWC6 A0A1E1WZG3 A0A1L8EDQ9 K7IQB4 A0A0A1WTX4 A0A182W258 A0A0A1X6Q9 A0A1B6H3K5 A0A1B6J3N2 A0A023FFB6 A0A182PK95 A0A182M3J2 A0A1B6HPL7 A0A1B6J5N7 A0A1B6FB04 A0A1B6JGA0 W8AVN9 B4PUA2 B3P6N6 A0A1I8M807 A0A182J6H5 A0A232F1M6 A0A224YZ84 A0A131YT23 A0A224YQT7 A0A131YDD2 B3LWJ5 B4QUR7 B4IJJ4 A0A1W4V8T7 Q9VBU6 Q296L6 B4GEZ8 A0A1I8NPS6 A0A140SRF8 U5EGZ4 E0VVP4 A0A0A9WKS1 A0A3B0KGW0 A0A131XPV6 A0A1B0ESB4 B4M5G1 A0A0K8VDM6 A0A034VJ28 A0A0M4EL41 A0A0A9ZEI7 B4JSL5 A0A0K8W1Q3
A0A3S2M9B6 A0A1W4X9J9 D6X1A4 A0A212EJL2 H9JBF4 A0A158NUW8 A0A195FPQ6 A0A3L8DFI4 A0A151WSV8 A0A336KQT1 A0A1W4X9B4 T1DF64 A0A151IRM5 A0A1Q3G3N1 A0A1Y1LLW8 A0A0K8TPZ6 Q16T78 A0A023EIL2 A0A182HD55 A0A1L8DUM1 A0A182RMQ7 A0A2M4AHY7 W5JML6 A0A182FVT0 A0A1J1INH3 T1E7W4 A0A2M3Z443 A0A182K8S4 A0A182QPE7 A0A154PEL8 A0A2M4AI32 A0A182N9Q2 A0A182YBQ4 F4X5Q3 A0A0L7QZ26 A0A2A3EN73 A0A0L0CIW1 A0A182VDM2 A0A182THS1 A0A182XFQ5 Q7PV51 A0A088ACJ9 A0A0M8ZX57 A0A195BVP6 V9II70 A0A067RF06 A0A0T6AWC6 A0A1E1WZG3 A0A1L8EDQ9 K7IQB4 A0A0A1WTX4 A0A182W258 A0A0A1X6Q9 A0A1B6H3K5 A0A1B6J3N2 A0A023FFB6 A0A182PK95 A0A182M3J2 A0A1B6HPL7 A0A1B6J5N7 A0A1B6FB04 A0A1B6JGA0 W8AVN9 B4PUA2 B3P6N6 A0A1I8M807 A0A182J6H5 A0A232F1M6 A0A224YZ84 A0A131YT23 A0A224YQT7 A0A131YDD2 B3LWJ5 B4QUR7 B4IJJ4 A0A1W4V8T7 Q9VBU6 Q296L6 B4GEZ8 A0A1I8NPS6 A0A140SRF8 U5EGZ4 E0VVP4 A0A0A9WKS1 A0A3B0KGW0 A0A131XPV6 A0A1B0ESB4 B4M5G1 A0A0K8VDM6 A0A034VJ28 A0A0M4EL41 A0A0A9ZEI7 B4JSL5 A0A0K8W1Q3
Pubmed
26354079
22651552
23622113
18362917
19820115
22118469
+ More
19121390 21347285 30249741 24330624 28004739 26369729 17510324 24945155 26483478 20920257 23761445 25244985 21719571 26108605 12364791 14747013 17210077 24845553 28503490 20075255 25830018 24495485 17994087 17550304 25315136 28648823 28797301 26830274 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20566863 25401762 28049606 25348373
19121390 21347285 30249741 24330624 28004739 26369729 17510324 24945155 26483478 20920257 23761445 25244985 21719571 26108605 12364791 14747013 17210077 24845553 28503490 20075255 25830018 24495485 17994087 17550304 25315136 28648823 28797301 26830274 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20566863 25401762 28049606 25348373
EMBL
GDQN01005547
JAT85507.1
NWSH01000315
PCG77518.1
KQ461198
KPJ06095.1
+ More
AK401462 KQ459606 BAM18084.1 KPI91493.1 ODYU01002544 SOQ40186.1 GAIX01006033 JAA86527.1 RSAL01000008 RVE53923.1 KQ971372 EFA10599.2 AGBW02014440 OWR41679.1 BABH01030843 BABH01030844 BABH01030845 ADTU01026803 KQ981335 KYN42575.1 QOIP01000009 RLU19066.1 KQ982769 KYQ50934.1 UFQS01000836 UFQS01001491 UFQT01000836 UFQT01001491 SSX07186.1 SSX11235.1 SSX27529.1 GALA01000757 JAA94095.1 KQ981122 KYN09349.1 GFDL01000635 JAV34410.1 GEZM01055817 JAV72925.1 GDAI01001392 JAI16211.1 CH477658 EAT37677.1 EJY57864.1 GAPW01004808 JAC08790.1 JXUM01033983 KQ561007 KXJ80064.1 GFDF01003954 JAV10130.1 GGFK01007082 MBW40403.1 ADMH02000674 ETN65371.1 CVRI01000056 CRL01791.1 GAMD01002697 JAA98893.1 GGFM01002512 MBW23263.1 AXCN02001803 KQ434874 KZC09660.1 GGFK01007125 MBW40446.1 GL888739 EGI58219.1 KQ414685 KOC63817.1 KZ288204 PBC33215.1 JRES01000335 KNC32191.1 AAAB01008986 EAA00447.4 KQ435840 KOX71389.1 KQ976401 KYM92652.1 JR048983 AEY60823.1 KK852510 KDR22352.1 LJIG01022648 KRT79449.1 GFAC01006997 JAT92191.1 GFDG01002005 JAV16794.1 GBXI01013404 GBXI01012322 GBXI01000210 JAD00888.1 JAD01970.1 JAD14082.1 GBXI01007696 JAD06596.1 GECZ01022029 GECZ01020757 GECZ01011509 GECZ01000549 JAS47740.1 JAS49012.1 JAS58260.1 JAS69220.1 GECU01029347 GECU01013925 GECU01003621 JAS78359.1 JAS93781.1 JAT04086.1 GBBK01003836 JAC20646.1 AXCM01001644 GECU01031079 JAS76627.1 GECU01036524 GECU01013201 JAS71182.1 JAS94505.1 GECZ01022458 GECZ01022125 JAS47311.1 JAS47644.1 GECU01031053 GECU01019451 GECU01009803 JAS76653.1 JAS88255.1 JAS97903.1 GAMC01017773 GAMC01017772 JAB88783.1 CM000160 EDW98789.1 CH954182 EDV53706.1 NNAY01001332 OXU24328.1 GFPF01008138 MAA19284.1 GEDV01007496 JAP81061.1 GFPF01008139 MAA19285.1 GEDV01011223 JAP77334.1 CH902617 EDV41589.1 CM000364 EDX14383.1 CH480848 EDW51185.1 AE014297 AY094863 AAF56431.1 AAM11216.1 CM000070 EAL28441.1 CH479182 EDW34183.1 ACL83566.2 GANO01003234 JAB56637.1 DS235813 EEB17450.1 GBHO01042363 GBHO01042360 GBHO01042358 GBHO01042355 GBHO01036393 GBHO01035593 GBHO01035592 GBHO01035590 GBHO01035589 GBHO01035588 GBHO01035583 GBHO01030522 GBHO01030520 GBHO01030516 GBHO01026043 GBHO01023778 GBHO01023774 GBHO01023772 GBHO01021346 GBHO01011284 GBHO01008523 GBHO01001830 GBHO01001829 GBHO01001827 GBRD01017221 GBRD01017220 GBRD01017219 GBRD01001193 GBRD01001192 GBRD01001191 JAG01241.1 JAG01244.1 JAG01246.1 JAG01249.1 JAG07211.1 JAG08011.1 JAG08012.1 JAG08014.1 JAG08015.1 JAG08016.1 JAG08021.1 JAG13082.1 JAG13084.1 JAG13088.1 JAG17561.1 JAG19826.1 JAG19830.1 JAG19832.1 JAG22258.1 JAG32320.1 JAG35081.1 JAG41774.1 JAG41775.1 JAG41777.1 JAG48606.1 OUUW01000008 SPP84331.1 GEFH01000189 JAP68392.1 AJWK01004548 CH940652 EDW59872.1 GDHF01030824 GDHF01020593 GDHF01015270 GDHF01008528 JAI21490.1 JAI31721.1 JAI37044.1 JAI43786.1 GAKP01017161 GAKP01017152 JAC41800.1 CP012526 ALC45940.1 GBHO01001831 JAG41773.1 CH916373 EDV94755.1 GDHF01030232 GDHF01017090 GDHF01007564 JAI22082.1 JAI35224.1 JAI44750.1
AK401462 KQ459606 BAM18084.1 KPI91493.1 ODYU01002544 SOQ40186.1 GAIX01006033 JAA86527.1 RSAL01000008 RVE53923.1 KQ971372 EFA10599.2 AGBW02014440 OWR41679.1 BABH01030843 BABH01030844 BABH01030845 ADTU01026803 KQ981335 KYN42575.1 QOIP01000009 RLU19066.1 KQ982769 KYQ50934.1 UFQS01000836 UFQS01001491 UFQT01000836 UFQT01001491 SSX07186.1 SSX11235.1 SSX27529.1 GALA01000757 JAA94095.1 KQ981122 KYN09349.1 GFDL01000635 JAV34410.1 GEZM01055817 JAV72925.1 GDAI01001392 JAI16211.1 CH477658 EAT37677.1 EJY57864.1 GAPW01004808 JAC08790.1 JXUM01033983 KQ561007 KXJ80064.1 GFDF01003954 JAV10130.1 GGFK01007082 MBW40403.1 ADMH02000674 ETN65371.1 CVRI01000056 CRL01791.1 GAMD01002697 JAA98893.1 GGFM01002512 MBW23263.1 AXCN02001803 KQ434874 KZC09660.1 GGFK01007125 MBW40446.1 GL888739 EGI58219.1 KQ414685 KOC63817.1 KZ288204 PBC33215.1 JRES01000335 KNC32191.1 AAAB01008986 EAA00447.4 KQ435840 KOX71389.1 KQ976401 KYM92652.1 JR048983 AEY60823.1 KK852510 KDR22352.1 LJIG01022648 KRT79449.1 GFAC01006997 JAT92191.1 GFDG01002005 JAV16794.1 GBXI01013404 GBXI01012322 GBXI01000210 JAD00888.1 JAD01970.1 JAD14082.1 GBXI01007696 JAD06596.1 GECZ01022029 GECZ01020757 GECZ01011509 GECZ01000549 JAS47740.1 JAS49012.1 JAS58260.1 JAS69220.1 GECU01029347 GECU01013925 GECU01003621 JAS78359.1 JAS93781.1 JAT04086.1 GBBK01003836 JAC20646.1 AXCM01001644 GECU01031079 JAS76627.1 GECU01036524 GECU01013201 JAS71182.1 JAS94505.1 GECZ01022458 GECZ01022125 JAS47311.1 JAS47644.1 GECU01031053 GECU01019451 GECU01009803 JAS76653.1 JAS88255.1 JAS97903.1 GAMC01017773 GAMC01017772 JAB88783.1 CM000160 EDW98789.1 CH954182 EDV53706.1 NNAY01001332 OXU24328.1 GFPF01008138 MAA19284.1 GEDV01007496 JAP81061.1 GFPF01008139 MAA19285.1 GEDV01011223 JAP77334.1 CH902617 EDV41589.1 CM000364 EDX14383.1 CH480848 EDW51185.1 AE014297 AY094863 AAF56431.1 AAM11216.1 CM000070 EAL28441.1 CH479182 EDW34183.1 ACL83566.2 GANO01003234 JAB56637.1 DS235813 EEB17450.1 GBHO01042363 GBHO01042360 GBHO01042358 GBHO01042355 GBHO01036393 GBHO01035593 GBHO01035592 GBHO01035590 GBHO01035589 GBHO01035588 GBHO01035583 GBHO01030522 GBHO01030520 GBHO01030516 GBHO01026043 GBHO01023778 GBHO01023774 GBHO01023772 GBHO01021346 GBHO01011284 GBHO01008523 GBHO01001830 GBHO01001829 GBHO01001827 GBRD01017221 GBRD01017220 GBRD01017219 GBRD01001193 GBRD01001192 GBRD01001191 JAG01241.1 JAG01244.1 JAG01246.1 JAG01249.1 JAG07211.1 JAG08011.1 JAG08012.1 JAG08014.1 JAG08015.1 JAG08016.1 JAG08021.1 JAG13082.1 JAG13084.1 JAG13088.1 JAG17561.1 JAG19826.1 JAG19830.1 JAG19832.1 JAG22258.1 JAG32320.1 JAG35081.1 JAG41774.1 JAG41775.1 JAG41777.1 JAG48606.1 OUUW01000008 SPP84331.1 GEFH01000189 JAP68392.1 AJWK01004548 CH940652 EDW59872.1 GDHF01030824 GDHF01020593 GDHF01015270 GDHF01008528 JAI21490.1 JAI31721.1 JAI37044.1 JAI43786.1 GAKP01017161 GAKP01017152 JAC41800.1 CP012526 ALC45940.1 GBHO01001831 JAG41773.1 CH916373 EDV94755.1 GDHF01030232 GDHF01017090 GDHF01007564 JAI22082.1 JAI35224.1 JAI44750.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000283053
UP000192223
UP000007266
+ More
UP000007151 UP000005204 UP000005205 UP000078541 UP000279307 UP000075809 UP000078492 UP000008820 UP000069940 UP000249989 UP000075900 UP000000673 UP000069272 UP000183832 UP000075881 UP000075886 UP000076502 UP000075884 UP000076408 UP000007755 UP000053825 UP000242457 UP000037069 UP000075903 UP000075902 UP000076407 UP000007062 UP000005203 UP000053105 UP000078540 UP000027135 UP000002358 UP000075920 UP000075885 UP000075883 UP000002282 UP000008711 UP000095301 UP000075880 UP000215335 UP000007801 UP000000304 UP000001292 UP000192221 UP000000803 UP000001819 UP000008744 UP000095300 UP000009046 UP000268350 UP000092461 UP000008792 UP000092553 UP000001070
UP000007151 UP000005204 UP000005205 UP000078541 UP000279307 UP000075809 UP000078492 UP000008820 UP000069940 UP000249989 UP000075900 UP000000673 UP000069272 UP000183832 UP000075881 UP000075886 UP000076502 UP000075884 UP000076408 UP000007755 UP000053825 UP000242457 UP000037069 UP000075903 UP000075902 UP000076407 UP000007062 UP000005203 UP000053105 UP000078540 UP000027135 UP000002358 UP000075920 UP000075885 UP000075883 UP000002282 UP000008711 UP000095301 UP000075880 UP000215335 UP000007801 UP000000304 UP000001292 UP000192221 UP000000803 UP000001819 UP000008744 UP000095300 UP000009046 UP000268350 UP000092461 UP000008792 UP000092553 UP000001070
Pfam
PF03248 Rer1
SUPFAM
SSF103473
SSF103473
ProteinModelPortal
A0A1E1WF01
A0A2A4K0S8
A0A194QMB3
I4DJJ4
A0A2H1VH83
S4PWX4
+ More
A0A3S2M9B6 A0A1W4X9J9 D6X1A4 A0A212EJL2 H9JBF4 A0A158NUW8 A0A195FPQ6 A0A3L8DFI4 A0A151WSV8 A0A336KQT1 A0A1W4X9B4 T1DF64 A0A151IRM5 A0A1Q3G3N1 A0A1Y1LLW8 A0A0K8TPZ6 Q16T78 A0A023EIL2 A0A182HD55 A0A1L8DUM1 A0A182RMQ7 A0A2M4AHY7 W5JML6 A0A182FVT0 A0A1J1INH3 T1E7W4 A0A2M3Z443 A0A182K8S4 A0A182QPE7 A0A154PEL8 A0A2M4AI32 A0A182N9Q2 A0A182YBQ4 F4X5Q3 A0A0L7QZ26 A0A2A3EN73 A0A0L0CIW1 A0A182VDM2 A0A182THS1 A0A182XFQ5 Q7PV51 A0A088ACJ9 A0A0M8ZX57 A0A195BVP6 V9II70 A0A067RF06 A0A0T6AWC6 A0A1E1WZG3 A0A1L8EDQ9 K7IQB4 A0A0A1WTX4 A0A182W258 A0A0A1X6Q9 A0A1B6H3K5 A0A1B6J3N2 A0A023FFB6 A0A182PK95 A0A182M3J2 A0A1B6HPL7 A0A1B6J5N7 A0A1B6FB04 A0A1B6JGA0 W8AVN9 B4PUA2 B3P6N6 A0A1I8M807 A0A182J6H5 A0A232F1M6 A0A224YZ84 A0A131YT23 A0A224YQT7 A0A131YDD2 B3LWJ5 B4QUR7 B4IJJ4 A0A1W4V8T7 Q9VBU6 Q296L6 B4GEZ8 A0A1I8NPS6 A0A140SRF8 U5EGZ4 E0VVP4 A0A0A9WKS1 A0A3B0KGW0 A0A131XPV6 A0A1B0ESB4 B4M5G1 A0A0K8VDM6 A0A034VJ28 A0A0M4EL41 A0A0A9ZEI7 B4JSL5 A0A0K8W1Q3
A0A3S2M9B6 A0A1W4X9J9 D6X1A4 A0A212EJL2 H9JBF4 A0A158NUW8 A0A195FPQ6 A0A3L8DFI4 A0A151WSV8 A0A336KQT1 A0A1W4X9B4 T1DF64 A0A151IRM5 A0A1Q3G3N1 A0A1Y1LLW8 A0A0K8TPZ6 Q16T78 A0A023EIL2 A0A182HD55 A0A1L8DUM1 A0A182RMQ7 A0A2M4AHY7 W5JML6 A0A182FVT0 A0A1J1INH3 T1E7W4 A0A2M3Z443 A0A182K8S4 A0A182QPE7 A0A154PEL8 A0A2M4AI32 A0A182N9Q2 A0A182YBQ4 F4X5Q3 A0A0L7QZ26 A0A2A3EN73 A0A0L0CIW1 A0A182VDM2 A0A182THS1 A0A182XFQ5 Q7PV51 A0A088ACJ9 A0A0M8ZX57 A0A195BVP6 V9II70 A0A067RF06 A0A0T6AWC6 A0A1E1WZG3 A0A1L8EDQ9 K7IQB4 A0A0A1WTX4 A0A182W258 A0A0A1X6Q9 A0A1B6H3K5 A0A1B6J3N2 A0A023FFB6 A0A182PK95 A0A182M3J2 A0A1B6HPL7 A0A1B6J5N7 A0A1B6FB04 A0A1B6JGA0 W8AVN9 B4PUA2 B3P6N6 A0A1I8M807 A0A182J6H5 A0A232F1M6 A0A224YZ84 A0A131YT23 A0A224YQT7 A0A131YDD2 B3LWJ5 B4QUR7 B4IJJ4 A0A1W4V8T7 Q9VBU6 Q296L6 B4GEZ8 A0A1I8NPS6 A0A140SRF8 U5EGZ4 E0VVP4 A0A0A9WKS1 A0A3B0KGW0 A0A131XPV6 A0A1B0ESB4 B4M5G1 A0A0K8VDM6 A0A034VJ28 A0A0M4EL41 A0A0A9ZEI7 B4JSL5 A0A0K8W1Q3
Ontologies
GO
PANTHER
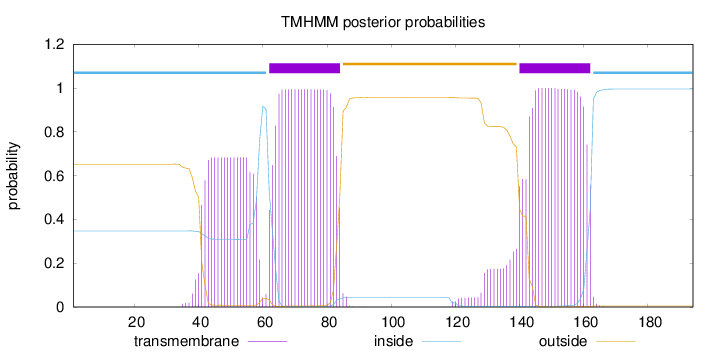
Topology
Length:
194
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
56.48944
Exp number, first 60 AAs:
12.28516
Total prob of N-in:
0.34749
POSSIBLE N-term signal
sequence
inside
1 - 61
TMhelix
62 - 84
outside
85 - 139
TMhelix
140 - 162
inside
163 - 194
Population Genetic Test Statistics
Pi
272.783201
Theta
188.424203
Tajima's D
0.829611
CLR
0.094311
CSRT
0.609319534023299
Interpretation
Uncertain
