Gene
KWMTBOMO05682
Annotation
PREDICTED:_uncharacterized_protein_LOC105391048_[Plutella_xylostella]
Full name
Galactose-1-phosphate uridylyltransferase
Location in the cell
Nuclear Reliability : 2.995
Sequence
CDS
ATGGCCAGTCAAGAAATACAGGAACAACTGCAACAACTCTTGGCTGCAAATGCAGCGCTACAAAAACAACTTGAAGGAAATGAGAGAGAAACAAAAGTTTGTAGAGTTACAGTCAAATTGCCTACATTTTGGGCTGACAAACCAGCTGTTTGGTTTGCTCAAATTGAAGCTCAATTCCAAATTGCCAATATTGTGACAGATACTACTAAATATAATTATGTTGCTCAAGCTATACTGGCAGCACAGGCTGACCTTGGGCTTGACAAATTGTCAGAACTAGCCGACAAAATTCTTGAGTTGGCCCAAACTCCTCCTATGGTATGTTCGACCGCAACATTTTCGAACAGTTTAACCCTTGGTTCATTAGCCAAACAACTAGAAGAGATCAGTAGCCAAGTTGCTGCCCTATCTAGGCGACAGAGCCGCCCTTATCGTGACAGATCATCATCTCGCGGCCCTAAAATTTGTTGGTATCATCGGAGAAATGAAAGGATAGTTGATGGGACCACAGGACTATCATCAATTGCCTCAACCACTGATATTGATCAACCAAGTGTTCGAGCAGTTGCATCTGGAAAGGTCATTTTTACTGAATTATTAGCAGAATTTCCAGACATTACACGACCACCAGGGCTGCCTCGAGTCGTTAAGCACCATACGGTCCATTTCATCAAAACAACGGATGGGCCTCCGATTGCCTGCCGTCCCCGCCGCTTAGCCCCTCAGAAACTAGTTGCAGCTAAGAAAGAGTTCGAGGATATGGTGCAATCTGGTACAGCTCGACCATCCAAAAGTGCCTGGTCATCACCACTGCATATGACAATTAAAAAAGACGACACCTGGCGTCCATGCGGCGATTACCGGGCCCTAAACGCCAGAACAGTTCCAGATAGGTACCCCGTGCGCCATATTCACGATTTTGCTCATAACCTCTCTGGTTCAAAAGTTTTCAGCACCATCGATTTAGTAAAAGCTTATCACCAGATACCGATCGCTGAGGAGGACGTGCCGAAGACAGCAATTGTTACACCGTTTGGCCTCTTCGAATTCCCATTCATGACTTTTGGGCTTCGAAACGCGGGACAGACATTCCAGCGATTTATTGATGAGGTCACACGGGGTCTTGATTTCTGTTTTCCCTACGTTGATGACATCCTCGTTCATTCAGCTGATGAGAATCTTCATCTGAAACACCTTCGAACCCTCTTCGAACGTCTACAAGAATACGGAGTTGTAATTAACCCCTCCAAATGTACTCTGGGAGTAGATGAAGTTAACTTCTTGGGCTATCACATAAACAGTGAAGGGACACGCCCTCCTCAGGCTCGCATCCAAGTCTTATTGGACTTTCCACCTCCTGAGACGGTACAAGGTATGCGCCGATTTCTCGGTATGCTGAACTACTACCGGCGTTTTGTTCCACATGCTGCTCAGTTTCAGGCACCGCTAATTGAGGCTCTCGTAGCAACAAAGAGCAAGGGTGCCAGGCCATTTCCTTGGTCCCCAGAGCTACTTCAGCAATTTGAAGCCTGCAAGCAAAGCTTGTCCTCGGCGACGCTATTACAACATCCTGTGAGTAATTCTAGTTTAGGTCTGTTCACGGATGCCTCCAGTGTTCATATTGGGTCTTGCCTTCAGCAAAAGGTAAATGGTAAGTGGTTTCCGCTATCTTTTTTTAGTAAGAAACTCACACCCCAGCAAAGACAGTGGCCGACTTACTACAGAGAATTATTAGCGGTATACGAAAGTGTTCAACATTTTCGATATATTTTAGAAGTCCAGCATGCCACAATTTTTACCGACCACAAACCACTCTTATATGCTTTCTTACAGAGACGCGAGAAGCTGCCACCTCCTCAGTTAAACCAGTTGTTTTTTATAAGCCAATTCACGACTGACATACAGTATATTAAGGGCGAGGACAATATCGTTGCTGACACAATGTCACGCATCGATATCAACGCCATCGCGCTGGATCAAGATCACGATGCACTCGCGAAGTCTCAGGCTACAGATTCTGAATTGAAAAATCTGTTACAAAATGGCTCTAGCCTGAGCCTAACTAAAATCAGCCCTCCAGAAGCGAAATCGGCAATTTATTGTGACGTTTCTACAGGTAAGCCACGTCCGTATTTGACCCTTTCCTTTCGAAGAACCGCGTTCAACCAACTGCACAACTTGAGTCATCCTGGTATCCGTGCTACCACACGCCTCGTGTCAGAACGTTTCGTTTGGCCATCGATGAAAAGGGACTGCCGTATTTGGTCTCGAAGCTGTGACGCATGTCAACGTAGCAAAGTTTCAAGACACAACTCAGCTCCTACTGGAAACTTTAACATACCATCCGGTCGCTTCATGCACGTGCATATAGACATTGTAGGGCCATTACCGGTTTGCAACGAATACCGATATTGTTTGACGGCAATTGATAGGGTTTCCAGGTGGCCAGAGGTGTGGCCTATGCGCACCATCACGGCAGAGGAAGTGGTGGAAACTTTTACCAGGGAGTGGATTTCCCGTTTTGGAGTACCGTCAATAATCACAACAGACCAAGGATCCCAATTTGAATCCGATTTGTTTAACCGGCTGCTGCTTCGATTCGCCACCAAGCGTATAAGGACTACCTCATATCATCCCTGCGCAAACGGAATGATTGAACGTGTCCATAGACAGCTCAAAGCGGCATTAATATGCCATCAAGATTCATGGGTAAGATCCCTTCCCATGGTTTTGCTGGGAATGCGGTCAGCCTTTAAAGAAGATTTGGAAGCCACAGTCGCAGAAACGGTGTACGGTGAACCTTTAAGACTCCTTGGAGAGCTGTTAATAGCTTCCTCGTCAATAGAAAAAATTGAGGATGCCACCGACTTTGTGGTACAATTGCGTAACAAAATGTCATCTATACGCCCAGTGCCTGCTTCTCGACATTGTAAAAACTCTCCATTTATATTCAAGGAATTGGAAAGCGCTTCTCATGTTTTCCTGAGAGACGATACCATCCGCAGGGCATTGCAGCCGCCATATACAGGTCCATATAGAGTATTAGAACGCTCCCCTGATGGGAAAACGTTCACACTGTCCATAAAAAATAACAAAGTTGTCGTGAGTGTAGACAGACTTAAGCCGGCTTTCATAGATCACATTGATGATGAACGGAAGCAAGCCGAGCCTTTGTCTTGTCAGCAAAATCAAGCCTTACGACCAATGGATCTGGAAAATCCGCCGCTGTTGCCGCTGCCAACAAGGCCGCTGTCAATTCCAATAAAAATTCAAGGTGAGAAAACAAAAGAGCCTTATACCACACGCTCAGGCAGACGTGTCAGATTTAAAAAAAATACTGATTTATAA
Protein
MASQEIQEQLQQLLAANAALQKQLEGNERETKVCRVTVKLPTFWADKPAVWFAQIEAQFQIANIVTDTTKYNYVAQAILAAQADLGLDKLSELADKILELAQTPPMVCSTATFSNSLTLGSLAKQLEEISSQVAALSRRQSRPYRDRSSSRGPKICWYHRRNERIVDGTTGLSSIASTTDIDQPSVRAVASGKVIFTELLAEFPDITRPPGLPRVVKHHTVHFIKTTDGPPIACRPRRLAPQKLVAAKKEFEDMVQSGTARPSKSAWSSPLHMTIKKDDTWRPCGDYRALNARTVPDRYPVRHIHDFAHNLSGSKVFSTIDLVKAYHQIPIAEEDVPKTAIVTPFGLFEFPFMTFGLRNAGQTFQRFIDEVTRGLDFCFPYVDDILVHSADENLHLKHLRTLFERLQEYGVVINPSKCTLGVDEVNFLGYHINSEGTRPPQARIQVLLDFPPPETVQGMRRFLGMLNYYRRFVPHAAQFQAPLIEALVATKSKGARPFPWSPELLQQFEACKQSLSSATLLQHPVSNSSLGLFTDASSVHIGSCLQQKVNGKWFPLSFFSKKLTPQQRQWPTYYRELLAVYESVQHFRYILEVQHATIFTDHKPLLYAFLQRREKLPPPQLNQLFFISQFTTDIQYIKGEDNIVADTMSRIDINAIALDQDHDALAKSQATDSELKNLLQNGSSLSLTKISPPEAKSAIYCDVSTGKPRPYLTLSFRRTAFNQLHNLSHPGIRATTRLVSERFVWPSMKRDCRIWSRSCDACQRSKVSRHNSAPTGNFNIPSGRFMHVHIDIVGPLPVCNEYRYCLTAIDRVSRWPEVWPMRTITAEEVVETFTREWISRFGVPSIITTDQGSQFESDLFNRLLLRFATKRIRTTSYHPCANGMIERVHRQLKAALICHQDSWVRSLPMVLLGMRSAFKEDLEATVAETVYGEPLRLLGELLIASSSIEKIEDATDFVVQLRNKMSSIRPVPASRHCKNSPFIFKELESASHVFLRDDTIRRALQPPYTGPYRVLERSPDGKTFTLSIKNNKVVVSVDRLKPAFIDHIDDERKQAEPLSCQQNQALRPMDLENPPLLPLPTRPLSIPIKIQGEKTKEPYTTRSGRRVRFKKNTDL
Summary
Catalytic Activity
alpha-D-galactose 1-phosphate + UDP-alpha-D-glucose = alpha-D-glucose 1-phosphate + UDP-alpha-D-galactose
Similarity
Belongs to the galactose-1-phosphate uridylyltransferase type 1 family.
Uniprot
A0A0J7KDT3
A0A0J7K7H4
A0A0J7MZD0
A0A0J7K6G5
A0A0J7K8C3
A0A0A1WEZ6
+ More
A0A0A1WJ40 A0A085NK58 A0A0A1XGK3 A0A147BLP1 A0A0J7K7B2 A0A085M208 A0A147BJG6 A0A0P4VHP4 A0A1Y1N1G2 A0A0K3CRM4 A0A2R5LDL5 A0A085N5A1 W4XB88 A7XDH3 D7GYM9 A0A0J7K7X5 J9KB96 A0A147BK98 A0A147BM81 A0A224Z7V0 A0A2R5L8Z1 A0A0J7NWH7 A0A147BIW8 A0A1Q3FTV9 A0A2G8JJ34 A0A147BJG9 A0A2M4A7J4 A0A2M4A7K1 W4YGV9 A0A147BTP0 A0A147BWJ6 K7ISS4 A0A0K8WJ70 A0A0J7K7I7 A0A0K8UV92 A0A0K8VH61 K7JCJ0 W4Y2D0 L7LZ63 Q4QQF8 A0A0J7K6W8 C7C1Z9 D6WTN9 A0A0A9Y7K2 A0A2S2NL30 A0A2L2YQU8 A0A1X7VKM7 A0A085N6P6 A0A085MR14 A0A131Z969 A0A085MTV7 A0A2L2YL91 Q4QQD4 Q6IFT8 C4QQP9 A0A085N6R2 A0A085MTF1 K7JCQ7 A0A085NC55 A0A131YP83 W4Y5A8 Q9BM79 J9K9L2 A0A0C9RQ48 A0A1Y1ME13 X1XU43 A0A085NRB3 X1XU40 Q9BM80 W4ZBK7 A0A1B6BX63 Q9BM81 A0A2G8JXA6 G7Y855 A0A0J7N8Y6 K7JAG6 A0A2W1BS51 W4XB89 A0A369THC0 G4LUJ9 Q6IFU3 X1XPJ2 X1X9N5 X1XTW4 A0A068XER4 K7JBH7 A0A267DVV7 A0A2S2PRR6 A0A085MZL4 A0A085MDY5
A0A0A1WJ40 A0A085NK58 A0A0A1XGK3 A0A147BLP1 A0A0J7K7B2 A0A085M208 A0A147BJG6 A0A0P4VHP4 A0A1Y1N1G2 A0A0K3CRM4 A0A2R5LDL5 A0A085N5A1 W4XB88 A7XDH3 D7GYM9 A0A0J7K7X5 J9KB96 A0A147BK98 A0A147BM81 A0A224Z7V0 A0A2R5L8Z1 A0A0J7NWH7 A0A147BIW8 A0A1Q3FTV9 A0A2G8JJ34 A0A147BJG9 A0A2M4A7J4 A0A2M4A7K1 W4YGV9 A0A147BTP0 A0A147BWJ6 K7ISS4 A0A0K8WJ70 A0A0J7K7I7 A0A0K8UV92 A0A0K8VH61 K7JCJ0 W4Y2D0 L7LZ63 Q4QQF8 A0A0J7K6W8 C7C1Z9 D6WTN9 A0A0A9Y7K2 A0A2S2NL30 A0A2L2YQU8 A0A1X7VKM7 A0A085N6P6 A0A085MR14 A0A131Z969 A0A085MTV7 A0A2L2YL91 Q4QQD4 Q6IFT8 C4QQP9 A0A085N6R2 A0A085MTF1 K7JCQ7 A0A085NC55 A0A131YP83 W4Y5A8 Q9BM79 J9K9L2 A0A0C9RQ48 A0A1Y1ME13 X1XU43 A0A085NRB3 X1XU40 Q9BM80 W4ZBK7 A0A1B6BX63 Q9BM81 A0A2G8JXA6 G7Y855 A0A0J7N8Y6 K7JAG6 A0A2W1BS51 W4XB89 A0A369THC0 G4LUJ9 Q6IFU3 X1XPJ2 X1X9N5 X1XTW4 A0A068XER4 K7JBH7 A0A267DVV7 A0A2S2PRR6 A0A085MZL4 A0A085MDY5
EC Number
2.7.7.12
Pubmed
EMBL
LBMM01009142
KMQ88359.1
LBMM01012479
KMQ86204.1
LBMM01013103
KMQ85815.1
+ More
LBMM01013040 KMQ85849.1 LBMM01012030 KMQ86466.1 GBXI01017046 JAC97245.1 GBXI01015889 JAC98402.1 KL367492 KFD69854.1 GBXI01003773 JAD10519.1 GEGO01003687 JAR91717.1 LBMM01012134 KMQ86388.1 KL363241 KFD51254.1 GEGO01004484 JAR90920.1 GDKW01003376 JAI53219.1 GEZM01017686 JAV90495.1 LN877232 CTR11708.1 GGLE01003443 MBY07569.1 KL367552 KFD64647.1 AAGJ04001742 EF426679 ABP48077.1 KQ973093 EFA13472.2 LBMM01012160 KMQ86374.1 ABLF02030693 ABLF02030697 GEGO01004649 JAR90755.1 GEGO01003506 JAR91898.1 GFPF01012375 MAA23521.1 GGLE01001814 MBY05940.1 LBMM01001181 KMQ96765.1 GEGO01004686 JAR90718.1 GFDL01003994 JAV31051.1 MRZV01001828 PIK35719.1 GEGO01004475 JAR90929.1 GGFK01003442 MBW36763.1 GGFK01003401 MBW36722.1 AAGJ04164195 AAGJ04164196 AAGJ04164197 GEGO01001281 JAR94123.1 GEGO01000286 JAR95118.1 AAZX01009606 GDHF01001116 JAI51198.1 LBMM01012567 KMQ86146.1 GDHF01021808 JAI30506.1 GDHF01014424 JAI37890.1 AAZX01006801 AAZX01023483 AAGJ04130731 GACK01008900 JAA56134.1 BN000785 CAJ00226.1 LBMM01012633 KMQ86092.1 FN356213 CAX83702.1 KQ971355 EFA07205.2 GBHO01015475 JAG28129.1 GGMR01005228 MBY17847.1 IAAA01040175 LAA10459.1 KL367544 KFD65142.1 KL367778 KFD59660.1 GEDV01001796 JAP86761.1 KL367655 KFD60653.1 IAAA01033850 LAA08869.1 BN000802 CAJ00250.1 BK004070 DAA04500.1 CABG01000050 CAZ37786.2 KL367543 KFD65158.1 KL367664 KFD60497.1 AAZX01004786 KL367519 KFD67051.1 GEDV01008182 JAP80375.1 AAGJ04112098 AY013569 AAK07487.1 ABLF02011086 ABLF02042879 GBYB01009311 JAG79078.1 GEZM01036621 JAV82565.1 ABLF02006352 ABLF02013150 ABLF02013854 ABLF02041505 KL367479 KFD72009.1 ABLF02040931 ABLF02040938 AY013565 AAK07486.1 AAGJ04000250 GEDC01031421 JAS05877.1 AY013563 AAK07485.1 MRZV01001127 MRZV01000556 PIK40396.1 PIK47817.1 DF142933 GAA49140.1 LBMM01008151 KMQ89105.1 AAZX01023279 KZ149973 PZC76007.1 QPMK01000035 RDD64024.1 CABG01000001 CCD58591.1 BK004066 DAA04495.1 ABLF02058233 ABLF02005645 ABLF02042063 AAZX01023264 NIVC01003188 PAA52757.1 GGMR01019415 MBY32034.1 KL367590 KFD62660.1 KL363200 KFD55431.1
LBMM01013040 KMQ85849.1 LBMM01012030 KMQ86466.1 GBXI01017046 JAC97245.1 GBXI01015889 JAC98402.1 KL367492 KFD69854.1 GBXI01003773 JAD10519.1 GEGO01003687 JAR91717.1 LBMM01012134 KMQ86388.1 KL363241 KFD51254.1 GEGO01004484 JAR90920.1 GDKW01003376 JAI53219.1 GEZM01017686 JAV90495.1 LN877232 CTR11708.1 GGLE01003443 MBY07569.1 KL367552 KFD64647.1 AAGJ04001742 EF426679 ABP48077.1 KQ973093 EFA13472.2 LBMM01012160 KMQ86374.1 ABLF02030693 ABLF02030697 GEGO01004649 JAR90755.1 GEGO01003506 JAR91898.1 GFPF01012375 MAA23521.1 GGLE01001814 MBY05940.1 LBMM01001181 KMQ96765.1 GEGO01004686 JAR90718.1 GFDL01003994 JAV31051.1 MRZV01001828 PIK35719.1 GEGO01004475 JAR90929.1 GGFK01003442 MBW36763.1 GGFK01003401 MBW36722.1 AAGJ04164195 AAGJ04164196 AAGJ04164197 GEGO01001281 JAR94123.1 GEGO01000286 JAR95118.1 AAZX01009606 GDHF01001116 JAI51198.1 LBMM01012567 KMQ86146.1 GDHF01021808 JAI30506.1 GDHF01014424 JAI37890.1 AAZX01006801 AAZX01023483 AAGJ04130731 GACK01008900 JAA56134.1 BN000785 CAJ00226.1 LBMM01012633 KMQ86092.1 FN356213 CAX83702.1 KQ971355 EFA07205.2 GBHO01015475 JAG28129.1 GGMR01005228 MBY17847.1 IAAA01040175 LAA10459.1 KL367544 KFD65142.1 KL367778 KFD59660.1 GEDV01001796 JAP86761.1 KL367655 KFD60653.1 IAAA01033850 LAA08869.1 BN000802 CAJ00250.1 BK004070 DAA04500.1 CABG01000050 CAZ37786.2 KL367543 KFD65158.1 KL367664 KFD60497.1 AAZX01004786 KL367519 KFD67051.1 GEDV01008182 JAP80375.1 AAGJ04112098 AY013569 AAK07487.1 ABLF02011086 ABLF02042879 GBYB01009311 JAG79078.1 GEZM01036621 JAV82565.1 ABLF02006352 ABLF02013150 ABLF02013854 ABLF02041505 KL367479 KFD72009.1 ABLF02040931 ABLF02040938 AY013565 AAK07486.1 AAGJ04000250 GEDC01031421 JAS05877.1 AY013563 AAK07485.1 MRZV01001127 MRZV01000556 PIK40396.1 PIK47817.1 DF142933 GAA49140.1 LBMM01008151 KMQ89105.1 AAZX01023279 KZ149973 PZC76007.1 QPMK01000035 RDD64024.1 CABG01000001 CCD58591.1 BK004066 DAA04495.1 ABLF02058233 ABLF02005645 ABLF02042063 AAZX01023264 NIVC01003188 PAA52757.1 GGMR01019415 MBY32034.1 KL367590 KFD62660.1 KL363200 KFD55431.1
Proteomes
PRIDE
Pfam
Interpro
IPR036397
RNaseH_sf
+ More
IPR041588 Integrase_H2C2
IPR000477 RT_dom
IPR012337 RNaseH-like_sf
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR001995 Peptidase_A2_cat
IPR021109 Peptidase_aspartic_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR034132 RP_Saci-like
IPR041373 RT_RNaseH
IPR005849 GalP_Utransf_N
IPR019779 GalP_UDPtransf1_His-AS
IPR001937 GalP_UDPtransf1
IPR036691 Endo/exonu/phosph_ase_sf
IPR036265 HIT-like_sf
IPR013032 EGF-like_CS
IPR027806 HARBI1_dom
IPR016024 ARM-type_fold
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR008383 API5
IPR038718 SNF2-like_sf
IPR027417 P-loop_NTPase
IPR000330 SNF2_N
IPR018061 Retropepsins
IPR041588 Integrase_H2C2
IPR000477 RT_dom
IPR012337 RNaseH-like_sf
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR001995 Peptidase_A2_cat
IPR021109 Peptidase_aspartic_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR034132 RP_Saci-like
IPR041373 RT_RNaseH
IPR005849 GalP_Utransf_N
IPR019779 GalP_UDPtransf1_His-AS
IPR001937 GalP_UDPtransf1
IPR036691 Endo/exonu/phosph_ase_sf
IPR036265 HIT-like_sf
IPR013032 EGF-like_CS
IPR027806 HARBI1_dom
IPR016024 ARM-type_fold
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR008383 API5
IPR038718 SNF2-like_sf
IPR027417 P-loop_NTPase
IPR000330 SNF2_N
IPR018061 Retropepsins
SUPFAM
ProteinModelPortal
A0A0J7KDT3
A0A0J7K7H4
A0A0J7MZD0
A0A0J7K6G5
A0A0J7K8C3
A0A0A1WEZ6
+ More
A0A0A1WJ40 A0A085NK58 A0A0A1XGK3 A0A147BLP1 A0A0J7K7B2 A0A085M208 A0A147BJG6 A0A0P4VHP4 A0A1Y1N1G2 A0A0K3CRM4 A0A2R5LDL5 A0A085N5A1 W4XB88 A7XDH3 D7GYM9 A0A0J7K7X5 J9KB96 A0A147BK98 A0A147BM81 A0A224Z7V0 A0A2R5L8Z1 A0A0J7NWH7 A0A147BIW8 A0A1Q3FTV9 A0A2G8JJ34 A0A147BJG9 A0A2M4A7J4 A0A2M4A7K1 W4YGV9 A0A147BTP0 A0A147BWJ6 K7ISS4 A0A0K8WJ70 A0A0J7K7I7 A0A0K8UV92 A0A0K8VH61 K7JCJ0 W4Y2D0 L7LZ63 Q4QQF8 A0A0J7K6W8 C7C1Z9 D6WTN9 A0A0A9Y7K2 A0A2S2NL30 A0A2L2YQU8 A0A1X7VKM7 A0A085N6P6 A0A085MR14 A0A131Z969 A0A085MTV7 A0A2L2YL91 Q4QQD4 Q6IFT8 C4QQP9 A0A085N6R2 A0A085MTF1 K7JCQ7 A0A085NC55 A0A131YP83 W4Y5A8 Q9BM79 J9K9L2 A0A0C9RQ48 A0A1Y1ME13 X1XU43 A0A085NRB3 X1XU40 Q9BM80 W4ZBK7 A0A1B6BX63 Q9BM81 A0A2G8JXA6 G7Y855 A0A0J7N8Y6 K7JAG6 A0A2W1BS51 W4XB89 A0A369THC0 G4LUJ9 Q6IFU3 X1XPJ2 X1X9N5 X1XTW4 A0A068XER4 K7JBH7 A0A267DVV7 A0A2S2PRR6 A0A085MZL4 A0A085MDY5
A0A0A1WJ40 A0A085NK58 A0A0A1XGK3 A0A147BLP1 A0A0J7K7B2 A0A085M208 A0A147BJG6 A0A0P4VHP4 A0A1Y1N1G2 A0A0K3CRM4 A0A2R5LDL5 A0A085N5A1 W4XB88 A7XDH3 D7GYM9 A0A0J7K7X5 J9KB96 A0A147BK98 A0A147BM81 A0A224Z7V0 A0A2R5L8Z1 A0A0J7NWH7 A0A147BIW8 A0A1Q3FTV9 A0A2G8JJ34 A0A147BJG9 A0A2M4A7J4 A0A2M4A7K1 W4YGV9 A0A147BTP0 A0A147BWJ6 K7ISS4 A0A0K8WJ70 A0A0J7K7I7 A0A0K8UV92 A0A0K8VH61 K7JCJ0 W4Y2D0 L7LZ63 Q4QQF8 A0A0J7K6W8 C7C1Z9 D6WTN9 A0A0A9Y7K2 A0A2S2NL30 A0A2L2YQU8 A0A1X7VKM7 A0A085N6P6 A0A085MR14 A0A131Z969 A0A085MTV7 A0A2L2YL91 Q4QQD4 Q6IFT8 C4QQP9 A0A085N6R2 A0A085MTF1 K7JCQ7 A0A085NC55 A0A131YP83 W4Y5A8 Q9BM79 J9K9L2 A0A0C9RQ48 A0A1Y1ME13 X1XU43 A0A085NRB3 X1XU40 Q9BM80 W4ZBK7 A0A1B6BX63 Q9BM81 A0A2G8JXA6 G7Y855 A0A0J7N8Y6 K7JAG6 A0A2W1BS51 W4XB89 A0A369THC0 G4LUJ9 Q6IFU3 X1XPJ2 X1X9N5 X1XTW4 A0A068XER4 K7JBH7 A0A267DVV7 A0A2S2PRR6 A0A085MZL4 A0A085MDY5
PDB
4OL8
E-value=1.46818e-65,
Score=638
Ontologies
KEGG
GO
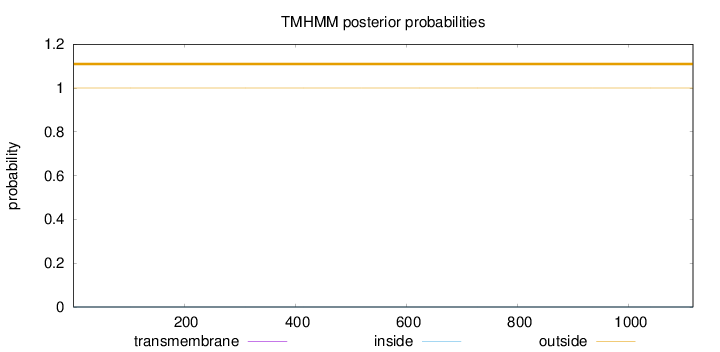
Topology
Length:
1115
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00219
Exp number, first 60 AAs:
0.00025
Total prob of N-in:
0.00012
outside
1 - 1115
Population Genetic Test Statistics
Pi
234.956035
Theta
27.118473
Tajima's D
0.732782
CLR
12.421425
CSRT
0.592270386480676
Interpretation
Uncertain
