Gene
KWMTBOMO05677
Pre Gene Modal
BGIBMGA006843
Annotation
PREDICTED:_protein_phosphatase_Slingshot_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.929
Sequence
CDS
ATGGCACTTGTCACAATTCGACGTTCTCCAAGTGTGCAAACACCAAGAAAATCGGATGAAGCAGATAAATCAAGTGACAAGGTCGAAGACAATGCCAACAACCGTGTCAACAAAAGCCTCAATGAATGCTACTTCGCGAATAAGGGCTCAGCAGTGGTGCTGGGGGGTACGGAGCGCGGCTGCATGGATCAGAGACCATCCCCAGTTCGCTCATGTCCCCAGTCTGACATACAAAATCACCTTCAATCTATGTTCTACTTATTGCGCCCTGAAGAGACGCTCAAAATGGCGGTGAAACTGGAAAGCGCTCACCCAGGCCGAACAAGATACCTGGTAGTGGTTTGCCGTAATGAAGAATCTGCGTTATTGGGTATCGACTGTAAGGAGCGCACAACTGTTGGACTAGTCCTAAGAGTCCTGGCCGACACATCCATAAAACTAGACGGAGATGGCGGATTCAGCGTCTGTGTATGTAACCAACAGCATATATTTAAGCCAGTCTCCGTACAAGCCATGTGGTCTGCTTTGCAAACGCTCCATCGGGCTAGTGCCAGAGCGCGACAGTTAAATCATTTTGCAGGGGGACCTTCGCATTCTTGGTGCGTCTTCTATGAGAATCATGTGGACTCTGATCGTTCTTGTCTTAACGAATGGCACGCTATGGATAACATCGAGTCGCGTCGACCGCCATCGCCTGATTCAGTCAGATTAAAGCCACGCGAAAGAGACGAAACAGAATGCGTAATAAGATGTACCCTCAAAGAAATCATGATGAGTGTGGATTTGGACGAAGTGACGAGTAAAGCCATCAGAGGTCGACTGGAGGAGGAGTTGGACATGGACCTCGCCGAGTACAAGTCATTCATTGACCACGAGATGCTCACGATATTAGGTCAGATGGACGCGCCGACAGAAATATTCGACCATGTTTATCTCGGTTCGGAATGGAACGCAAGTAACCTCGAAGAATTACAGAGAAACGGAGTCAGACACATCTTAAACGTTACAAGGGAGATAGACAATTTCTTTCCCGGAATGTTTGAGTATCTGAACATAAGGGTTTACGACGACGACAAGACTGACCTATTGAAACATTGGGACAACACGTTTAAGTATATAAATAAAGCGAGAAATGAAGGTTCAAAAGTACTCGTGCACTGTAAAATGGGCATCAGCAGATCCGCCTCTGTCGTAATAGCTTACGCAATGAAAGCATTCAATTGGAATTTCGACAGAGCCCTGAAGCACGTTAAAGCGAAAAGGAGTTGTATCAAACCGAACACGAATTTTTTGAACCAACTAGAGACTTATCAAGGTATCCTTGATGCAATGAAAAATAAAGAGAAATTACAAAGATCAAAGTCTGAAACGAATTTAAAATTGCCTAAATCGGTTACGAAAAATGAGAACAAATCAATGGAGCCGACGCCTCTAGTCTTAGCTTTGACGGGATCGTACAGCGGCCGGCCGCGCTCGTGGTCGCCGGACACCAAGATAGCGTCTCAGCTCCTCCCTGCCCTCCCGCCGACGTCGGTTTCCCTCGAGAATTTAGCCTCCGAGACCAGGCACATGCTAATGCCGTGTGCCAACGGAAGCTACAGCGTATCACCGAATCAAATAATCAGATTAAAAGGCGAAGGCGCTCCATCAGTGAAACACATTGTGAATGAAATCGAGAACGCTACGTCGGGCGATCGCAGGGATCTGAGTCGTAGATATCAAAAATTGATTTTTGGTAGTCAGTATGATTGTCTCAGCGATCGCTCTGTCGATGTTCCTAGCGCTCCGAGCGTTCCAACGCAAACTTCTCCGACGCGCAGTTCCAGTCTCAAATATGCGCAGCATTATTTCGAAGGAGATAGAATACACACGTGGGACCCCGGGGAAACGTCTGCGTGTCGAGACGAGATCGGTCGCAATCTTAGTAATAGTGATAATTTAGTAAAAAGTGACAGTGGCATCGTAGATATAAAAACGAATAAAGTGACAATTAGTGACGGTTACAGTTTTATCAATTTGACAGAACGGAATTTAGAAAGTGAAGGTAAACGAGTTCCCGACGAAGAAGCAGCACCTCCGAGTCGACAAAGTTCGTGGAGTTCATTCGATAGCGCAGTTGTACTCGATCTGTCTCGGCACTCTTCGTGGGGCTCGTACGATACGAGAGGCGCAAAAACGCAGATCATCGGTCGGGAGGAATCTATTAAACGAGCCAAAGATCGATCTGACGATCGCAACTTTAAGAAGGATGATACAGAACGCGGTGCCGAATGCGCGGCCGCCTCACAAAAACAGCTCTCCGATCTCAGCATCATAAGAGAACACAACGAGGCACGCAACGCCGAGCGGCGACTCACCGACAGATCCGGCCTCGTGGCGGACAATGAAAGAAAATTTAACGAGACCTGTGCCATATTAAAAGAACTCGCCAACGCCGCAGCCAGAATGGAAAAAGCGAGAGACCGAGGGGCGTCCACGTGGAGCGGGCGCCTGGCCGCCTCCGTGCCCGTCGACACGTGGCTGCGAGCCGGCCTACGGCGCCGCCGGCCTGTCTGTGCTTCGCAAGGCGACTTGCCGCGCGCCGCAGCCGGACTCGTCAGTAACCTCAAAAAGGAGTTCGAAGCTCGCTCCGAAACCGATACATTGCGACGGTCGGAACCTCGCACTCGGACGGCAGCCAACGAGCACGCCGATAGATCTCCGGCGTCGCCACCTTCCGGCGAAGATCTCTCCGTGAAAGTCTTAGTAGACAGATACGACAGGCCCGGGCGTTCGAGGTCGGAGTCGGTCGGAACGAAAGGTCCACAAGAGTCCGTCTCGAAGAAATGTCGGTTGACGGCGGAGAGCGAGAGTCGCACTCGCACTCGCAGCGGCGCGGTGGCGCTGCGGAACTCGTTCTGCGGCGCCTTCCGCGGGCTCGGCGCCGACCGGCCGCCGCCCGCCCCCACCGTCCTCGCGTTGGCTCCTATCGACTATAACGATGTGGTGGTATCTACTGTGATGTCAAAAGCTCAAAACAAAAAACAATTGCAACACGGGAAGACGCATCCGTTAACGAGGCTCAACCTAAATCGGTCAAACAATAGATGTTCAAATCCTGTATATAACACTATGTAG
Protein
MALVTIRRSPSVQTPRKSDEADKSSDKVEDNANNRVNKSLNECYFANKGSAVVLGGTERGCMDQRPSPVRSCPQSDIQNHLQSMFYLLRPEETLKMAVKLESAHPGRTRYLVVVCRNEESALLGIDCKERTTVGLVLRVLADTSIKLDGDGGFSVCVCNQQHIFKPVSVQAMWSALQTLHRASARARQLNHFAGGPSHSWCVFYENHVDSDRSCLNEWHAMDNIESRRPPSPDSVRLKPRERDETECVIRCTLKEIMMSVDLDEVTSKAIRGRLEEELDMDLAEYKSFIDHEMLTILGQMDAPTEIFDHVYLGSEWNASNLEELQRNGVRHILNVTREIDNFFPGMFEYLNIRVYDDDKTDLLKHWDNTFKYINKARNEGSKVLVHCKMGISRSASVVIAYAMKAFNWNFDRALKHVKAKRSCIKPNTNFLNQLETYQGILDAMKNKEKLQRSKSETNLKLPKSVTKNENKSMEPTPLVLALTGSYSGRPRSWSPDTKIASQLLPALPPTSVSLENLASETRHMLMPCANGSYSVSPNQIIRLKGEGAPSVKHIVNEIENATSGDRRDLSRRYQKLIFGSQYDCLSDRSVDVPSAPSVPTQTSPTRSSSLKYAQHYFEGDRIHTWDPGETSACRDEIGRNLSNSDNLVKSDSGIVDIKTNKVTISDGYSFINLTERNLESEGKRVPDEEAAPPSRQSSWSSFDSAVVLDLSRHSSWGSYDTRGAKTQIIGREESIKRAKDRSDDRNFKKDDTERGAECAAASQKQLSDLSIIREHNEARNAERRLTDRSGLVADNERKFNETCAILKELANAAARMEKARDRGASTWSGRLAASVPVDTWLRAGLRRRRPVCASQGDLPRAAAGLVSNLKKEFEARSETDTLRRSEPRTRTAANEHADRSPASPPSGEDLSVKVLVDRYDRPGRSRSESVGTKGPQESVSKKCRLTAESESRTRTRSGAVALRNSFCGAFRGLGADRPPPAPTVLALAPIDYNDVVVSTVMSKAQNKKQLQHGKTHPLTRLNLNRSNNRCSNPVYNTM
Summary
Uniprot
EMBL
ODYU01005623
SOQ46658.1
KQ459606
KPI91502.1
AGBW02014631
OWR41293.1
+ More
NWSH01002534 PCG68050.1 KQ434785 KZC05023.1 GEDC01030369 JAS06929.1 KQ971372 KYB25369.1 KQ977513 KYN02195.1 GECZ01002412 JAS67357.1 GEDC01025115 JAS12183.1 GECZ01013826 JAS55943.1 KQ979480 KYN21251.1 DS235132 EEB12395.1 GFXV01002304 MBW14109.1 GFDF01000997 JAV13087.1 GFDF01000996 JAV13088.1
NWSH01002534 PCG68050.1 KQ434785 KZC05023.1 GEDC01030369 JAS06929.1 KQ971372 KYB25369.1 KQ977513 KYN02195.1 GECZ01002412 JAS67357.1 GEDC01025115 JAS12183.1 GECZ01013826 JAS55943.1 KQ979480 KYN21251.1 DS235132 EEB12395.1 GFXV01002304 MBW14109.1 GFDF01000997 JAV13087.1 GFDF01000996 JAV13088.1
Proteomes
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
CDD
ProteinModelPortal
PDB
2NT2
E-value=1.28152e-56,
Score=560
Ontologies
GO
Topology
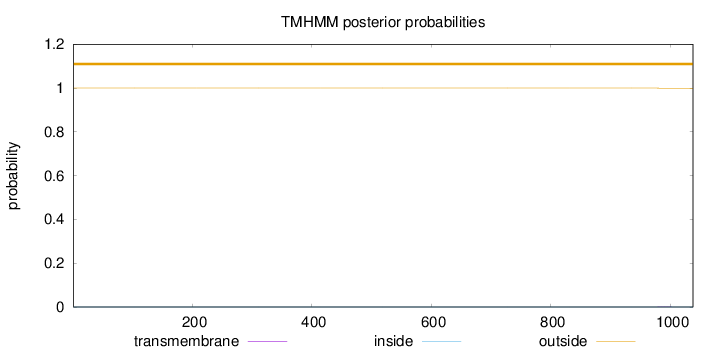
Length:
1038
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01205
Exp number, first 60 AAs:
0.00036
Total prob of N-in:
0.00012
outside
1 - 1038
Population Genetic Test Statistics
Pi
223.617159
Theta
194.933873
Tajima's D
0.423971
CLR
0.370892
CSRT
0.496775161241938
Interpretation
Uncertain
