Gene
KWMTBOMO05673 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006841
Annotation
PREDICTED:_A-kinase_anchor_protein_1?_mitochondrial_isoform_X2_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.466
Sequence
CDS
ATGGCACCTTGTCGTCAACTACTCATGTGGTCAGTACCATCCATAGCAGTCTTATTGGGTATTTTTTGGTTCAAGAAAAAAAGGGAATATGCAAAGTCTGACCCAGGTGGAAGAGAACGGCTAAAGAGTTTGAAAGAGGAACTTGCTGAGGTTTTGAACGCACAAGCTGAGGCAAATCGAGGCTCGCCTATAAGCAAATTAGAGCACTCTATCATTAAATCATCTCCTATTGATATCATACCTAACGGCACAAATTCACAGCGATCATCCCCTTTGGAACTCACTGACGAAGAAATTGATCTGGAGATAGAAAAAATAATCAGAATAAAATCCATTGGAAAAGAGAAGAGAATGTCAGCTGGCTTTGACAAACAAAATTTGATTGTGAGCTCATGTACTACAGAACCCTCCTTCGTAGTTAAGGAGTTGCAGAATAATTTGTGTTCACATGTTCAGCTCCAACAATCTTCACCTAAAAACTCAGAAATGCACATAATAGACAGTAATCATAGCTTTAGTAGTAATGTAAATTCTAATAAGGATCTGCTTGATTCAGAAGCAGAATGTTATAAAAAAAATAGTACAAACAATGGGACTGAGACCTTTGTGTCTGAAGGTGATACAATAGAAAATACCTCTGAAGGTACAGAAGGATTACAACCACTTGATGTTGTTGATGAACCCTGCAATACAGGTGACAACAATGATAGGTTTTCCACTTCACAAAGAAGACTCCTTTCGGAGAGGGACTCTGCTAATCATAGTCCTGTTGATCCTTTATTAGCTAGTCCCTCAATGTGCAATTTTTCAGATAATCACAGTGAGGGTTCAAGTGACAGTGGTAAAGGCTGTTCTGAGGCTGCTAGCCCTCCTCCTGCCAGTTTAAATATAGTTCCAACAGAAACAGACCTTAGGATACACCAATTTGTTTTATCTCAAAATCTTGTAGGTCTTTTAATAGGAAAACATGGTTCATTTGTGACTCAAATTAAAGCCAAAACTGGTGCAAGTGTTTACGTCAGAAGGCATCCTGAATCTTCCAAACAGAAAATATGTGCAGTTGAAGGAACACAAAGTGAAATAGAAGCAGCTTTAGATATGATAAAAGAAAAATTTCCTGAAAAAAAATTCCCAAACTTCTCACTGCAAGAAATCAGTGCTGAACTCTACCAAAAACTCACACCTCTCATCCCTGAGTTTTTACAATTGGTCGAATCTGTGAATAACGACACGATATTGACGTGTCTGGTGAGTGCGGGGCACTTCTTCCTCCAGCAGCCCCTGCACCCCACGTTTCCGTCGCTGCACGTGCTTCACCGGCTAATGGCCGCCACGTACCAGAACCCGGATGTGCCGTCGTTGCCACGCCCCGTGAAAGAGGGAGCTATCTGTGCTGCGCCTACTGAAAATAATTGGTATCGCGCCCAAGTGATATCTACCTCTGAAGAGAATGATACATCTGTCGTGAAACTAGTGGACTTCGGTGGATATCTCACTGTTGACAACGATCAGCTCAAACAGATCCGCTCCGATTTTATGACACTGCCTTTCCAGGCGACGGAGGCTCTCTTAGCATTTGTGAAACCAGCTAACAATGTAGAGTGGAGCGGAGAGGCTTTACGTATAATGGCCGGTTTGACCGCGGGGCAGCTCCTACACGCGCAGGTCGCAGGCTACGACGAGACTGGCCTGCCTCTTGTACATCTATACCTCACATTACATCCTCAACAAGTCATATTCCTGAATAGAGAGCTAGTAGAACGCGGACTGGCGGAGTGGGATGTTCCACCAGACTCCTGA
Protein
MAPCRQLLMWSVPSIAVLLGIFWFKKKREYAKSDPGGRERLKSLKEELAEVLNAQAEANRGSPISKLEHSIIKSSPIDIIPNGTNSQRSSPLELTDEEIDLEIEKIIRIKSIGKEKRMSAGFDKQNLIVSSCTTEPSFVVKELQNNLCSHVQLQQSSPKNSEMHIIDSNHSFSSNVNSNKDLLDSEAECYKKNSTNNGTETFVSEGDTIENTSEGTEGLQPLDVVDEPCNTGDNNDRFSTSQRRLLSERDSANHSPVDPLLASPSMCNFSDNHSEGSSDSGKGCSEAASPPPASLNIVPTETDLRIHQFVLSQNLVGLLIGKHGSFVTQIKAKTGASVYVRRHPESSKQKICAVEGTQSEIEAALDMIKEKFPEKKFPNFSLQEISAELYQKLTPLIPEFLQLVESVNNDTILTCLVSAGHFFLQQPLHPTFPSLHVLHRLMAATYQNPDVPSLPRPVKEGAICAAPTENNWYRAQVISTSEENDTSVVKLVDFGGYLTVDNDQLKQIRSDFMTLPFQATEALLAFVKPANNVEWSGEALRIMAGLTAGQLLHAQVAGYDETGLPLVHLYLTLHPQQVIFLNRELVERGLAEWDVPPDS
Summary
Uniprot
EMBL
BABH01030817
NWSH01005003
PCG64505.1
ODYU01009959
SOQ54721.1
GDQN01001159
+ More
GDQN01000571 JAT89895.1 JAT90483.1 GDQN01009487 GDQN01009221 JAT81567.1 JAT81833.1 KQ459606 KPI91509.1 KQ461198 KPJ06110.1 GAIX01003817 JAA88743.1 AGBW02014631 OWR41298.1 JTDY01000002 KOB79527.1 GDQN01004999 GDQN01002689 JAT86055.1 JAT88365.1
GDQN01000571 JAT89895.1 JAT90483.1 GDQN01009487 GDQN01009221 JAT81567.1 JAT81833.1 KQ459606 KPI91509.1 KQ461198 KPJ06110.1 GAIX01003817 JAA88743.1 AGBW02014631 OWR41298.1 JTDY01000002 KOB79527.1 GDQN01004999 GDQN01002689 JAT86055.1 JAT88365.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF54791
SSF54791
Gene 3D
ProteinModelPortal
PDB
5J39
E-value=1.67901e-11,
Score=169
Ontologies
GO
PANTHER
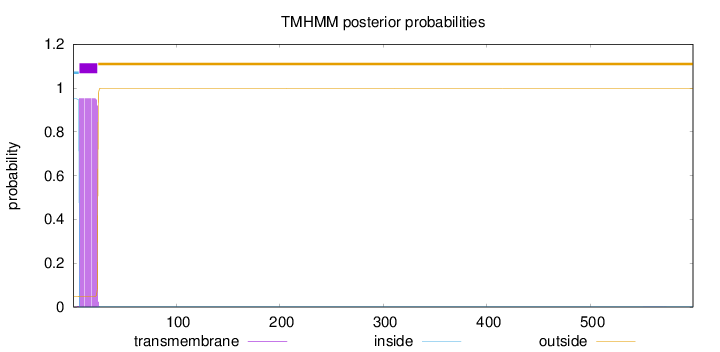
Topology
Length:
599
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
17.154
Exp number, first 60 AAs:
17.1482
Total prob of N-in:
0.95177
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 24
outside
25 - 599
Population Genetic Test Statistics
Pi
158.009861
Theta
15.267451
Tajima's D
-0.223273
CLR
14.464953
CSRT
0.306884655767212
Interpretation
Uncertain
