Pre Gene Modal
BGIBMGA014046
Annotation
putative_ribosomal_RNA_methyltransferase_[Danaus_plexippus]
Full name
Putative rRNA methyltransferase
Alternative Name
2'-O-ribose RNA methyltransferase SPB1 homolog
Location in the cell
Nuclear Reliability : 4.001
Sequence
CDS
ATGGGAAAGAAAACAAAAGTAGGAAAACAACGGAAAGATAAATACTACCAACTAGCTAAAGAAACAGGATATAGATCAAGAGCTGCATTCAAATTAATACAATTGAACAGGAAATTTGGATTTTTGCAGAAATCACGAGTATGTATCGATTTATGTGCTGCACCTGGTGGGTGGATGCAGGTGGCCCACCAAAATATGCCAGTCTCGAGCATAGTAATAGGAGTCGATTTGTTTCCAATAAAACCAGTTCCAGGTTGTATAGGTCTGACTGAAGATATCACTACAGAAAAATGTAAGGCAGCAATCAAAAAAGAAATAAAAACATGGAAAGCTGATGTGGTACTTAATGACGGTGCACCCAATGTCGGTCTTAATTGGATCCATGATGCCTATCAACAAGCTTGTCTAACGTTGAGTGCTTTAAAACTTGCAACGAACTTCCTCAGAGAAGGTGGTTGGTTTGTGACTAAAGTATTCAGATCGAAAGACTATCATGCACTTTTATGGGTTCTGAAACAATTCTTCAAAAAAGTACATGCTACAAAGCCCCAAGCTTCTCGTAATGAGTCTTCTGAAATTTTCGTTGTATGTCAGGGCTATATTGCACCAGATACAATTGATCCTAAATTTCTCGACCCCAAACATGTCTTTGAAGATCTTGATATAGTTAAAAAACAGCATATAAATATATTACATCCTGAGAAACAAAAAAAAGCAAAAGCAGAAGGATATAAAGAAAATGATTATACCACACATCATAAAGTAAAACTGTCTGATTTTTTGGGAAAAGATGATCCAGTAGATCTACTTCATGGTTGCTCTGAGATTGTTTTCGATGATAAGGAAATAGAAACTCATCCAAAGACAACACCAGAAATAAAGGAATGTTGTAAGGATATTAAGGTGCTTGGACGAAAAGACTTAAAATCACTGCTGTCATGGTTCAAGCAAATGAAAGAATATAAATCGTCTAAGGAGAAAAAAGATGATGAGCCCCAAACCAATGATGAAAACGAAGAACATGAAACAAATGAGGATGCTGATACTGAAGAAGGTGATGTGGACACACAAATTGCAGAACTGCAGGATGAAGAGAAGCGACAATTGAAACGTAAAAGGAAGCAGTTGAATAAACAGAGACAAAAACTAGCTGAAAAAATGAATCTTAAGATGGTATTGAAAGGAGATGATGGTCCTATTTTGGAAAGTAATGATATGTTTCAACTCTCTGATATCAAGAATGCAGAGGAACTGAACACGCTTATAGATCAGCAGCCTGACACCGTGGTTGATGGAGATGATGCTTTTGATGATGAGATTCAACTTAAGCCAAAGTACTTGAAATACGATGTTGAGGAAACCAAGCTCGATTCATCAGGACTTTATTACAAAGATTCAGACAGTGAAATTGAAATGGAATCTGATTCAGAATCTGAACAGGAAAAAGAGTCATTAGCCTTCTCAGACTCAGACACTGAATCGAAAAAGATCAAAAAATTAACTAAATTGAATACTCTAAAAAACAGTAAAGTGAGTCCAGCAAACAAATTGAATAGATTACAGTCTATTCCAAAGAATAACCCGTTGCTCACTGATCTGGACAACACTGACCCAGTTTCGAAAAGGACTATGAAAGCTCAGCTATGGTTCGAGAAAGATTCTTTTAAAAATATTGATGAAGAAGCTGATGAAGGTATAGATTTGGACAAGTTAACAGAAACATTCAAGGAAAAAGGGAAGAAAGTCATTGAAGATGTGGAGTCAAGTAAACAAACCAGTGCACTAAAAAGAAAATTGTCAGACAACGAGAGTTCAGATGATGATTCCTCTGACAGTGATTATGATGTTGAAGAAAACGTTACTCCCAGTGCTTCTAGTGCTGGGCCAAAAGCCGGCAAAGAAGGTTTTGAAGTTGTCTCCCAAGATCCAGAACTGAAAAAACTAAAGAAAACTTTGAAAATGGACGCAGAGACACTTGCTTTAGGAACCATGATAGCTACATCAAAAAAATTCAAGAGGGATTTAATTGATGACGCGTGGAATAGATACGCGTTTAACGATAAAAACTTACCAGATTGGTTTATGGAAGATGAAAAGAAACATATGAAGAAAGACATTGTGATACCGGAGAAGTATACTGATGAATACAAGAATAGATTACAAGACATCAATGTGAGGCCTATCAAGAAGGTGATTGAAGCCAAAGCGCGGAAGAAGAAGCGCTTGATCAAAAAGATGGAACGTGCAAAGAAAAAAGTCGAGGCGGTAATGGAGAACGCTGATATGTCCGAGCGAGAGAAAGCACAACAAGTGAGACAACTTTACAAGAAGGCACAATCTGATGGTAAAAAGAAAATCACTTATGTGGTGGCCAAGAAACATACAACTGCTAAGCGTATGAAGAGACCACAGGGCGTTAAGGGCCAGTACAAAGTGGTAGATCCGCGCATGAAAAAGGACTTACGTGCCCAAAAGGCAAAGGAGAAAACAAAAGGTCGAGGTAAAAAACAACCCAAGAAACAAAACAAGCCTAAAAAAGTCCCCAAAAAGAAAGGAAAAGCAAAGTGA
Protein
MGKKTKVGKQRKDKYYQLAKETGYRSRAAFKLIQLNRKFGFLQKSRVCIDLCAAPGGWMQVAHQNMPVSSIVIGVDLFPIKPVPGCIGLTEDITTEKCKAAIKKEIKTWKADVVLNDGAPNVGLNWIHDAYQQACLTLSALKLATNFLREGGWFVTKVFRSKDYHALLWVLKQFFKKVHATKPQASRNESSEIFVVCQGYIAPDTIDPKFLDPKHVFEDLDIVKKQHINILHPEKQKKAKAEGYKENDYTTHHKVKLSDFLGKDDPVDLLHGCSEIVFDDKEIETHPKTTPEIKECCKDIKVLGRKDLKSLLSWFKQMKEYKSSKEKKDDEPQTNDENEEHETNEDADTEEGDVDTQIAELQDEEKRQLKRKRKQLNKQRQKLAEKMNLKMVLKGDDGPILESNDMFQLSDIKNAEELNTLIDQQPDTVVDGDDAFDDEIQLKPKYLKYDVEETKLDSSGLYYKDSDSEIEMESDSESEQEKESLAFSDSDTESKKIKKLTKLNTLKNSKVSPANKLNRLQSIPKNNPLLTDLDNTDPVSKRTMKAQLWFEKDSFKNIDEEADEGIDLDKLTETFKEKGKKVIEDVESSKQTSALKRKLSDNESSDDDSSDSDYDVEENVTPSASSAGPKAGKEGFEVVSQDPELKKLKKTLKMDAETLALGTMIATSKKFKRDLIDDAWNRYAFNDKNLPDWFMEDEKKHMKKDIVIPEKYTDEYKNRLQDINVRPIKKVIEAKARKKKRLIKKMERAKKKVEAVMENADMSEREKAQQVRQLYKKAQSDGKKKITYVVAKKHTTAKRMKRPQGVKGQYKVVDPRMKKDLRAQKAKEKTKGRGKKQPKKQNKPKKVPKKKGKAK
Summary
Description
Probable methyltransferase involved in the maturation of rRNA and in the biogenesis of ribosomal subunits.
Catalytic Activity
a ribonucleotide in rRNA + S-adenosyl-L-methionine = a 2'-O-methylribonucleotide in rRNA + H(+) + S-adenosyl-L-homocysteine
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. RNA methyltransferase RlmE family. SPB1 subfamily.
Feature
chain Putative rRNA methyltransferase
Uniprot
H9JWY1
A0A2H1VBU3
A0A2A4JNH1
A0A194QRF2
S4PCI3
A0A3S2NAS2
+ More
A0A212ERR9 A0A194PE31 A0A154PNE2 A0A158NJW7 A0A067QZB0 E2BWY3 F4WIM5 N6TQ08 A0A195BHM4 D6X2B8 A0A195DW47 A0A026WQ43 A0A195F561 E9IMG1 A0A0L7R2X0 A0A151XE37 A0A2J7R304 E2AKJ6 A0A232FCH2 K7J012 A0A0C9QN46 A0A0C9R253 A0A088A7G6 A0A0J7KYH7 A0A151I7Y1 A0A0M8ZSQ5 A0A2A3EB54 A0A0T6B5J2 A0A310SM55 A0A1Y1MEY5 A0A1B0DPK2 A0A034WPD8 J9K0Y7 A0A1L8DX33 A0A1L8DX35 A0A0A1WMR3 A0A0N7Z9H4 A0A0K8V4M7 W8C434 A0A1A9X8R9 A0A0V0G7Y1 A0A1B0C3B3 Q17ME6 B4MA57 A0A1A9WT14 T1HXG8 A0A1A9ZEH1 A0A1Q3F1M3 B0WJS3 A0A1A9UQC1 U5EYF1 B4L6Y5 A0A1W4VGE3 A0A336L2X8 B4IMG9 Q9VXK5 A0A1Q3F1N3 A0A336MNM4 B3NXF9 A0A0Q5TK67 B4PXA9 B4JX39 A0A2M3Z119 A0A2M3ZG57 A0A182SGY5 A0A182RNS4 A0A1D2N9L0 A0A2M4A492 A0A2M4A450 A0A2M4A4X9 A0A1B3PD75 A0A182II28 A0A1B0GA10 Q7QD87 A0A2S2R6S3 A0A182G625 A0A084VC43
A0A212ERR9 A0A194PE31 A0A154PNE2 A0A158NJW7 A0A067QZB0 E2BWY3 F4WIM5 N6TQ08 A0A195BHM4 D6X2B8 A0A195DW47 A0A026WQ43 A0A195F561 E9IMG1 A0A0L7R2X0 A0A151XE37 A0A2J7R304 E2AKJ6 A0A232FCH2 K7J012 A0A0C9QN46 A0A0C9R253 A0A088A7G6 A0A0J7KYH7 A0A151I7Y1 A0A0M8ZSQ5 A0A2A3EB54 A0A0T6B5J2 A0A310SM55 A0A1Y1MEY5 A0A1B0DPK2 A0A034WPD8 J9K0Y7 A0A1L8DX33 A0A1L8DX35 A0A0A1WMR3 A0A0N7Z9H4 A0A0K8V4M7 W8C434 A0A1A9X8R9 A0A0V0G7Y1 A0A1B0C3B3 Q17ME6 B4MA57 A0A1A9WT14 T1HXG8 A0A1A9ZEH1 A0A1Q3F1M3 B0WJS3 A0A1A9UQC1 U5EYF1 B4L6Y5 A0A1W4VGE3 A0A336L2X8 B4IMG9 Q9VXK5 A0A1Q3F1N3 A0A336MNM4 B3NXF9 A0A0Q5TK67 B4PXA9 B4JX39 A0A2M3Z119 A0A2M3ZG57 A0A182SGY5 A0A182RNS4 A0A1D2N9L0 A0A2M4A492 A0A2M4A450 A0A2M4A4X9 A0A1B3PD75 A0A182II28 A0A1B0GA10 Q7QD87 A0A2S2R6S3 A0A182G625 A0A084VC43
EC Number
2.1.1.-
Pubmed
19121390
26354079
23622113
22118469
21347285
24845553
+ More
20798317 21719571 23537049 18362917 19820115 24508170 30249741 21282665 28648823 20075255 28004739 25348373 25830018 27129103 24495485 17510324 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27289101 12364791 14747013 17210077 26483478 24438588
20798317 21719571 23537049 18362917 19820115 24508170 30249741 21282665 28648823 20075255 28004739 25348373 25830018 27129103 24495485 17510324 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27289101 12364791 14747013 17210077 26483478 24438588
EMBL
BABH01041553
ODYU01001529
SOQ37872.1
NWSH01001013
PCG73134.1
KQ461198
+ More
KPJ06116.1 GAIX01004191 JAA88369.1 RSAL01000312 RVE42665.1 AGBW02012957 OWR44159.1 KQ459606 KPI91522.1 KQ434978 KZC12984.1 ADTU01018446 KK853080 KDR11689.1 GL451188 EFN79847.1 GL888176 EGI65961.1 APGK01056102 KB741274 KB632130 ENN71340.1 ERL88981.1 KQ976465 KYM84305.1 KQ971371 EFA09884.1 KQ980304 KYN16784.1 KK107135 QOIP01000006 EZA58088.1 RLU21317.1 KQ981820 KYN35312.1 GL764146 EFZ18245.1 KQ414664 KOC65178.1 KQ982254 KYQ58625.1 NEVH01007824 PNF35206.1 GL440281 EFN66047.1 NNAY01000463 OXU28190.1 GBYB01002077 JAG71844.1 GBYB01002080 JAG71847.1 LBMM01002043 KMQ95359.1 KQ978390 KYM94299.1 KQ435918 KOX68749.1 KZ288296 PBC28920.1 LJIG01009668 KRT82620.1 KQ759878 OAD62242.1 GEZM01033160 JAV84389.1 AJVK01008144 GAKP01001501 JAC57451.1 ABLF02040185 GFDF01003071 JAV11013.1 GFDF01003067 JAV11017.1 GBXI01013993 JAD00299.1 GDKW01000539 JAI56056.1 GDHF01018460 JAI33854.1 GAMC01005769 GAMC01005767 JAC00787.1 GECL01002027 JAP04097.1 JXJN01024889 CH477206 EAT47892.1 CH940655 EDW66116.1 ACPB03008619 GFDL01013590 JAV21455.1 DS231963 EDS29416.1 GANO01000338 JAB59533.1 CH933812 EDW06131.1 UFQS01001510 UFQT01001510 SSX11313.1 SSX30881.1 CH480996 EDW46314.1 AE014298 AY069518 AAF48557.1 AAL39663.1 GFDL01013583 JAV21462.1 UFQT01001823 SSX31906.1 CH954180 EDV46918.1 KQS30246.1 CM000162 EDX01872.1 CH916376 EDV95315.1 GGFM01001454 MBW22205.1 GGFM01006714 MBW27465.1 LJIJ01000133 ODN01931.1 GGFK01002229 MBW35550.1 GGFK01002265 MBW35586.1 GGFK01002494 MBW35815.1 KU659873 AOG17672.1 APCN01000610 CCAG010018793 AAAB01008859 EAA07890.5 GGMS01016197 MBY85400.1 JXUM01147113 KQ570300 KXJ68392.1 ATLV01010166 KE524565 KFB35537.1
KPJ06116.1 GAIX01004191 JAA88369.1 RSAL01000312 RVE42665.1 AGBW02012957 OWR44159.1 KQ459606 KPI91522.1 KQ434978 KZC12984.1 ADTU01018446 KK853080 KDR11689.1 GL451188 EFN79847.1 GL888176 EGI65961.1 APGK01056102 KB741274 KB632130 ENN71340.1 ERL88981.1 KQ976465 KYM84305.1 KQ971371 EFA09884.1 KQ980304 KYN16784.1 KK107135 QOIP01000006 EZA58088.1 RLU21317.1 KQ981820 KYN35312.1 GL764146 EFZ18245.1 KQ414664 KOC65178.1 KQ982254 KYQ58625.1 NEVH01007824 PNF35206.1 GL440281 EFN66047.1 NNAY01000463 OXU28190.1 GBYB01002077 JAG71844.1 GBYB01002080 JAG71847.1 LBMM01002043 KMQ95359.1 KQ978390 KYM94299.1 KQ435918 KOX68749.1 KZ288296 PBC28920.1 LJIG01009668 KRT82620.1 KQ759878 OAD62242.1 GEZM01033160 JAV84389.1 AJVK01008144 GAKP01001501 JAC57451.1 ABLF02040185 GFDF01003071 JAV11013.1 GFDF01003067 JAV11017.1 GBXI01013993 JAD00299.1 GDKW01000539 JAI56056.1 GDHF01018460 JAI33854.1 GAMC01005769 GAMC01005767 JAC00787.1 GECL01002027 JAP04097.1 JXJN01024889 CH477206 EAT47892.1 CH940655 EDW66116.1 ACPB03008619 GFDL01013590 JAV21455.1 DS231963 EDS29416.1 GANO01000338 JAB59533.1 CH933812 EDW06131.1 UFQS01001510 UFQT01001510 SSX11313.1 SSX30881.1 CH480996 EDW46314.1 AE014298 AY069518 AAF48557.1 AAL39663.1 GFDL01013583 JAV21462.1 UFQT01001823 SSX31906.1 CH954180 EDV46918.1 KQS30246.1 CM000162 EDX01872.1 CH916376 EDV95315.1 GGFM01001454 MBW22205.1 GGFM01006714 MBW27465.1 LJIJ01000133 ODN01931.1 GGFK01002229 MBW35550.1 GGFK01002265 MBW35586.1 GGFK01002494 MBW35815.1 KU659873 AOG17672.1 APCN01000610 CCAG010018793 AAAB01008859 EAA07890.5 GGMS01016197 MBY85400.1 JXUM01147113 KQ570300 KXJ68392.1 ATLV01010166 KE524565 KFB35537.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000283053
UP000007151
UP000053268
+ More
UP000076502 UP000005205 UP000027135 UP000008237 UP000007755 UP000019118 UP000030742 UP000078540 UP000007266 UP000078492 UP000053097 UP000279307 UP000078541 UP000053825 UP000075809 UP000235965 UP000000311 UP000215335 UP000002358 UP000005203 UP000036403 UP000078542 UP000053105 UP000242457 UP000092462 UP000007819 UP000092443 UP000092460 UP000008820 UP000008792 UP000091820 UP000015103 UP000092445 UP000002320 UP000078200 UP000009192 UP000192221 UP000001292 UP000000803 UP000008711 UP000002282 UP000001070 UP000075901 UP000075900 UP000094527 UP000075840 UP000092444 UP000007062 UP000069940 UP000249989 UP000030765
UP000076502 UP000005205 UP000027135 UP000008237 UP000007755 UP000019118 UP000030742 UP000078540 UP000007266 UP000078492 UP000053097 UP000279307 UP000078541 UP000053825 UP000075809 UP000235965 UP000000311 UP000215335 UP000002358 UP000005203 UP000036403 UP000078542 UP000053105 UP000242457 UP000092462 UP000007819 UP000092443 UP000092460 UP000008820 UP000008792 UP000091820 UP000015103 UP000092445 UP000002320 UP000078200 UP000009192 UP000192221 UP000001292 UP000000803 UP000008711 UP000002282 UP000001070 UP000075901 UP000075900 UP000094527 UP000075840 UP000092444 UP000007062 UP000069940 UP000249989 UP000030765
Pfam
Interpro
IPR029063
SAM-dependent_MTases
+ More
IPR001128 Cyt_P450
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR012920 rRNA_MeTfrase_Spb1_C
IPR015507 rRNA-MeTfrase_E
IPR036396 Cyt_P450_sf
IPR028589 Spb1-like
IPR024576 rRNA_MeTfrase_Spb1_DUF3381
IPR032501 Prot_ATP_ID_OB
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR005937 26S_Psome_P45-like
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR004776 Mem_trans
IPR020846 MFS_dom
IPR001128 Cyt_P450
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR012920 rRNA_MeTfrase_Spb1_C
IPR015507 rRNA-MeTfrase_E
IPR036396 Cyt_P450_sf
IPR028589 Spb1-like
IPR024576 rRNA_MeTfrase_Spb1_DUF3381
IPR032501 Prot_ATP_ID_OB
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR005937 26S_Psome_P45-like
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR004776 Mem_trans
IPR020846 MFS_dom
Gene 3D
CDD
ProteinModelPortal
H9JWY1
A0A2H1VBU3
A0A2A4JNH1
A0A194QRF2
S4PCI3
A0A3S2NAS2
+ More
A0A212ERR9 A0A194PE31 A0A154PNE2 A0A158NJW7 A0A067QZB0 E2BWY3 F4WIM5 N6TQ08 A0A195BHM4 D6X2B8 A0A195DW47 A0A026WQ43 A0A195F561 E9IMG1 A0A0L7R2X0 A0A151XE37 A0A2J7R304 E2AKJ6 A0A232FCH2 K7J012 A0A0C9QN46 A0A0C9R253 A0A088A7G6 A0A0J7KYH7 A0A151I7Y1 A0A0M8ZSQ5 A0A2A3EB54 A0A0T6B5J2 A0A310SM55 A0A1Y1MEY5 A0A1B0DPK2 A0A034WPD8 J9K0Y7 A0A1L8DX33 A0A1L8DX35 A0A0A1WMR3 A0A0N7Z9H4 A0A0K8V4M7 W8C434 A0A1A9X8R9 A0A0V0G7Y1 A0A1B0C3B3 Q17ME6 B4MA57 A0A1A9WT14 T1HXG8 A0A1A9ZEH1 A0A1Q3F1M3 B0WJS3 A0A1A9UQC1 U5EYF1 B4L6Y5 A0A1W4VGE3 A0A336L2X8 B4IMG9 Q9VXK5 A0A1Q3F1N3 A0A336MNM4 B3NXF9 A0A0Q5TK67 B4PXA9 B4JX39 A0A2M3Z119 A0A2M3ZG57 A0A182SGY5 A0A182RNS4 A0A1D2N9L0 A0A2M4A492 A0A2M4A450 A0A2M4A4X9 A0A1B3PD75 A0A182II28 A0A1B0GA10 Q7QD87 A0A2S2R6S3 A0A182G625 A0A084VC43
A0A212ERR9 A0A194PE31 A0A154PNE2 A0A158NJW7 A0A067QZB0 E2BWY3 F4WIM5 N6TQ08 A0A195BHM4 D6X2B8 A0A195DW47 A0A026WQ43 A0A195F561 E9IMG1 A0A0L7R2X0 A0A151XE37 A0A2J7R304 E2AKJ6 A0A232FCH2 K7J012 A0A0C9QN46 A0A0C9R253 A0A088A7G6 A0A0J7KYH7 A0A151I7Y1 A0A0M8ZSQ5 A0A2A3EB54 A0A0T6B5J2 A0A310SM55 A0A1Y1MEY5 A0A1B0DPK2 A0A034WPD8 J9K0Y7 A0A1L8DX33 A0A1L8DX35 A0A0A1WMR3 A0A0N7Z9H4 A0A0K8V4M7 W8C434 A0A1A9X8R9 A0A0V0G7Y1 A0A1B0C3B3 Q17ME6 B4MA57 A0A1A9WT14 T1HXG8 A0A1A9ZEH1 A0A1Q3F1M3 B0WJS3 A0A1A9UQC1 U5EYF1 B4L6Y5 A0A1W4VGE3 A0A336L2X8 B4IMG9 Q9VXK5 A0A1Q3F1N3 A0A336MNM4 B3NXF9 A0A0Q5TK67 B4PXA9 B4JX39 A0A2M3Z119 A0A2M3ZG57 A0A182SGY5 A0A182RNS4 A0A1D2N9L0 A0A2M4A492 A0A2M4A450 A0A2M4A4X9 A0A1B3PD75 A0A182II28 A0A1B0GA10 Q7QD87 A0A2S2R6S3 A0A182G625 A0A084VC43
PDB
6EM5
E-value=6.62098e-75,
Score=717
Ontologies
GO
GO:0000453
GO:0005506
GO:0016705
GO:0020037
GO:0005730
GO:0008649
GO:0005634
GO:0008168
GO:0001510
GO:0016435
GO:0031167
GO:0008650
GO:0030687
GO:0000463
GO:0000466
GO:0030163
GO:0036402
GO:0008233
GO:0005737
GO:0005524
GO:0051301
GO:0055085
GO:0016021
GO:0006364
GO:0032259
GO:0008270
GO:0003700
GO:0043565
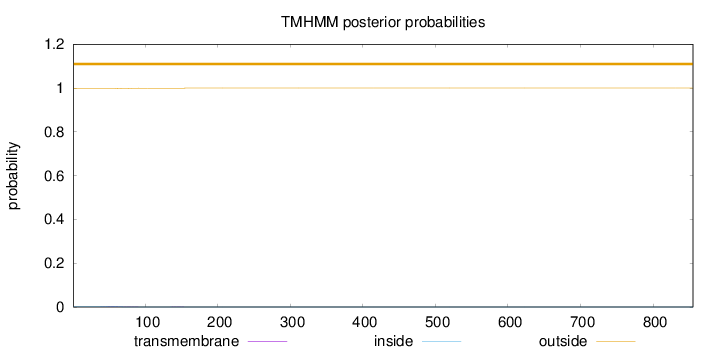
Topology
Subcellular location
Length:
855
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0536800000000001
Exp number, first 60 AAs:
0.02263
Total prob of N-in:
0.00293
outside
1 - 855
Population Genetic Test Statistics
Pi
234.487055
Theta
203.875447
Tajima's D
0.615012
CLR
0.418173
CSRT
0.553122343882806
Interpretation
Uncertain
